
Roseovarius faecimaris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseovarius
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
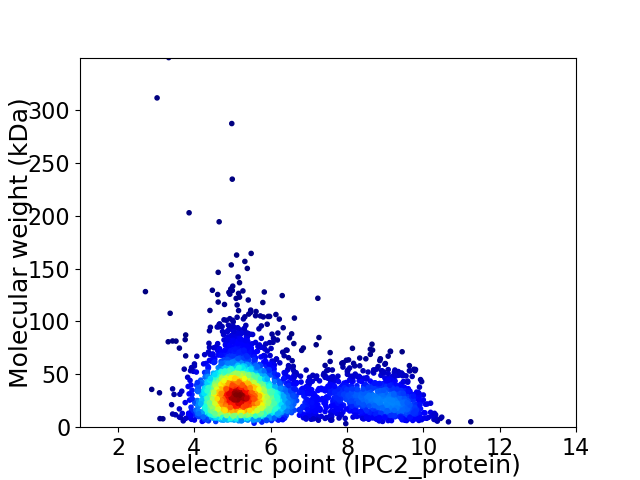
Virtual 2D-PAGE plot for 3738 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I6IQ10|A0A6I6IQ10_9RHOB 6-phosphogluconolactonase OS=Roseovarius faecimaris OX=2494550 GN=pgl PE=3 SV=1
MM1 pKa = 8.12SIMTVSSNEE10 pKa = 3.51QLMIEE15 pKa = 4.49LVNSFRR21 pKa = 11.84LDD23 pKa = 3.52PYY25 pKa = 11.2GEE27 pKa = 3.99AVRR30 pKa = 11.84IGLTSQEE37 pKa = 3.83IAALGLPQGSASPLAGTYY55 pKa = 7.47TLHH58 pKa = 6.17VAAGQHH64 pKa = 5.25SSWMIANDD72 pKa = 3.87DD73 pKa = 4.13FSHH76 pKa = 6.38TGAGGSSAGQRR87 pKa = 11.84MEE89 pKa = 3.84DD90 pKa = 2.93AGYY93 pKa = 10.09VFSGSYY99 pKa = 9.6HH100 pKa = 5.43WGEE103 pKa = 4.24NIGYY107 pKa = 8.23RR108 pKa = 11.84ASSGTLDD115 pKa = 3.34TTQAIYY121 pKa = 10.6HH122 pKa = 6.58LNRR125 pKa = 11.84ALILSDD131 pKa = 2.77GHH133 pKa = 6.67RR134 pKa = 11.84ANLLNPVFRR143 pKa = 11.84EE144 pKa = 4.03VGIGQANGDD153 pKa = 3.91YY154 pKa = 10.59LGYY157 pKa = 9.92DD158 pKa = 3.06ATMITQNFATSNGATFLTGVVYY180 pKa = 11.09NDD182 pKa = 3.15INGDD186 pKa = 3.53DD187 pKa = 4.37FYY189 pKa = 11.79SVGEE193 pKa = 4.17GVGGVTFSLSGKK205 pKa = 10.34SDD207 pKa = 3.26TTYY210 pKa = 11.12GAGGYY215 pKa = 7.25SVKK218 pKa = 9.39VTSGGTQTVTIAYY231 pKa = 6.71GTTEE235 pKa = 3.46ISAAVHH241 pKa = 6.08IGSEE245 pKa = 4.33NVKK248 pKa = 10.5LDD250 pKa = 4.85LIDD253 pKa = 3.63EE254 pKa = 4.57TVLRR258 pKa = 11.84SSSSLTLLSGATDD271 pKa = 3.36AVLLGLEE278 pKa = 4.4DD279 pKa = 3.75LSLYY283 pKa = 10.79GNSRR287 pKa = 11.84HH288 pKa = 6.06NHH290 pKa = 4.48LTGNAGDD297 pKa = 3.79NMLAGGLGNDD307 pKa = 4.64TIDD310 pKa = 3.65GGAGVDD316 pKa = 3.55TVKK319 pKa = 10.69INAGSSEE326 pKa = 4.19VTVEE330 pKa = 3.81ISGNDD335 pKa = 3.46IIVTSAEE342 pKa = 3.94GTDD345 pKa = 3.38VYY347 pKa = 11.39RR348 pKa = 11.84NVEE351 pKa = 4.06FFSFSNQTLTSSEE364 pKa = 3.91VRR366 pKa = 11.84EE367 pKa = 4.21LDD369 pKa = 3.47PSNSTTSSNATSPGPSSSTTGLFIAGGDD397 pKa = 3.92GADD400 pKa = 4.53LLDD403 pKa = 3.87GSNYY407 pKa = 9.92NDD409 pKa = 3.65SLKK412 pKa = 11.04GGNGNDD418 pKa = 3.51TLSGYY423 pKa = 10.86GGDD426 pKa = 4.23DD427 pKa = 3.57NLPGGEE433 pKa = 4.73GDD435 pKa = 4.01DD436 pKa = 3.89QVSGGAGNDD445 pKa = 3.67LLGGGLGDD453 pKa = 3.82DD454 pKa = 4.37TLNAGAGNDD463 pKa = 3.44QGGGGEE469 pKa = 4.38GNDD472 pKa = 3.73VINAGEE478 pKa = 4.17GDD480 pKa = 4.47DD481 pKa = 4.26IFSGGAGNDD490 pKa = 3.52LVQGADD496 pKa = 3.27GHH498 pKa = 6.64DD499 pKa = 3.48WLAASYY505 pKa = 11.61GNDD508 pKa = 3.25TVTGGAGNDD517 pKa = 3.91TIGAGHH523 pKa = 6.05GHH525 pKa = 6.62DD526 pKa = 5.09RR527 pKa = 11.84INGGAGDD534 pKa = 4.04DD535 pKa = 3.63QIGAGNHH542 pKa = 7.12DD543 pKa = 4.26DD544 pKa = 3.97TLLGGDD550 pKa = 4.13GNDD553 pKa = 3.93FLGGGYY559 pKa = 10.45GRR561 pKa = 11.84DD562 pKa = 3.72VLDD565 pKa = 4.69GEE567 pKa = 5.04AGQDD571 pKa = 3.7TLNGGYY577 pKa = 10.64GSDD580 pKa = 3.37TLTGGVGQDD589 pKa = 2.75EE590 pKa = 4.92FVFNTLSGNGNDD602 pKa = 4.27VITDD606 pKa = 3.8FQVGIDD612 pKa = 3.83HH613 pKa = 6.88IKK615 pKa = 10.83LVGLDD620 pKa = 3.69LGHH623 pKa = 6.97NPMQNLDD630 pKa = 3.42IRR632 pKa = 11.84DD633 pKa = 3.85TANGAVIEE641 pKa = 4.43YY642 pKa = 10.49GDD644 pKa = 3.66FTITVSGVSSHH655 pKa = 6.63SLTADD660 pKa = 3.26DD661 pKa = 6.7FIFF664 pKa = 4.65
MM1 pKa = 8.12SIMTVSSNEE10 pKa = 3.51QLMIEE15 pKa = 4.49LVNSFRR21 pKa = 11.84LDD23 pKa = 3.52PYY25 pKa = 11.2GEE27 pKa = 3.99AVRR30 pKa = 11.84IGLTSQEE37 pKa = 3.83IAALGLPQGSASPLAGTYY55 pKa = 7.47TLHH58 pKa = 6.17VAAGQHH64 pKa = 5.25SSWMIANDD72 pKa = 3.87DD73 pKa = 4.13FSHH76 pKa = 6.38TGAGGSSAGQRR87 pKa = 11.84MEE89 pKa = 3.84DD90 pKa = 2.93AGYY93 pKa = 10.09VFSGSYY99 pKa = 9.6HH100 pKa = 5.43WGEE103 pKa = 4.24NIGYY107 pKa = 8.23RR108 pKa = 11.84ASSGTLDD115 pKa = 3.34TTQAIYY121 pKa = 10.6HH122 pKa = 6.58LNRR125 pKa = 11.84ALILSDD131 pKa = 2.77GHH133 pKa = 6.67RR134 pKa = 11.84ANLLNPVFRR143 pKa = 11.84EE144 pKa = 4.03VGIGQANGDD153 pKa = 3.91YY154 pKa = 10.59LGYY157 pKa = 9.92DD158 pKa = 3.06ATMITQNFATSNGATFLTGVVYY180 pKa = 11.09NDD182 pKa = 3.15INGDD186 pKa = 3.53DD187 pKa = 4.37FYY189 pKa = 11.79SVGEE193 pKa = 4.17GVGGVTFSLSGKK205 pKa = 10.34SDD207 pKa = 3.26TTYY210 pKa = 11.12GAGGYY215 pKa = 7.25SVKK218 pKa = 9.39VTSGGTQTVTIAYY231 pKa = 6.71GTTEE235 pKa = 3.46ISAAVHH241 pKa = 6.08IGSEE245 pKa = 4.33NVKK248 pKa = 10.5LDD250 pKa = 4.85LIDD253 pKa = 3.63EE254 pKa = 4.57TVLRR258 pKa = 11.84SSSSLTLLSGATDD271 pKa = 3.36AVLLGLEE278 pKa = 4.4DD279 pKa = 3.75LSLYY283 pKa = 10.79GNSRR287 pKa = 11.84HH288 pKa = 6.06NHH290 pKa = 4.48LTGNAGDD297 pKa = 3.79NMLAGGLGNDD307 pKa = 4.64TIDD310 pKa = 3.65GGAGVDD316 pKa = 3.55TVKK319 pKa = 10.69INAGSSEE326 pKa = 4.19VTVEE330 pKa = 3.81ISGNDD335 pKa = 3.46IIVTSAEE342 pKa = 3.94GTDD345 pKa = 3.38VYY347 pKa = 11.39RR348 pKa = 11.84NVEE351 pKa = 4.06FFSFSNQTLTSSEE364 pKa = 3.91VRR366 pKa = 11.84EE367 pKa = 4.21LDD369 pKa = 3.47PSNSTTSSNATSPGPSSSTTGLFIAGGDD397 pKa = 3.92GADD400 pKa = 4.53LLDD403 pKa = 3.87GSNYY407 pKa = 9.92NDD409 pKa = 3.65SLKK412 pKa = 11.04GGNGNDD418 pKa = 3.51TLSGYY423 pKa = 10.86GGDD426 pKa = 4.23DD427 pKa = 3.57NLPGGEE433 pKa = 4.73GDD435 pKa = 4.01DD436 pKa = 3.89QVSGGAGNDD445 pKa = 3.67LLGGGLGDD453 pKa = 3.82DD454 pKa = 4.37TLNAGAGNDD463 pKa = 3.44QGGGGEE469 pKa = 4.38GNDD472 pKa = 3.73VINAGEE478 pKa = 4.17GDD480 pKa = 4.47DD481 pKa = 4.26IFSGGAGNDD490 pKa = 3.52LVQGADD496 pKa = 3.27GHH498 pKa = 6.64DD499 pKa = 3.48WLAASYY505 pKa = 11.61GNDD508 pKa = 3.25TVTGGAGNDD517 pKa = 3.91TIGAGHH523 pKa = 6.05GHH525 pKa = 6.62DD526 pKa = 5.09RR527 pKa = 11.84INGGAGDD534 pKa = 4.04DD535 pKa = 3.63QIGAGNHH542 pKa = 7.12DD543 pKa = 4.26DD544 pKa = 3.97TLLGGDD550 pKa = 4.13GNDD553 pKa = 3.93FLGGGYY559 pKa = 10.45GRR561 pKa = 11.84DD562 pKa = 3.72VLDD565 pKa = 4.69GEE567 pKa = 5.04AGQDD571 pKa = 3.7TLNGGYY577 pKa = 10.64GSDD580 pKa = 3.37TLTGGVGQDD589 pKa = 2.75EE590 pKa = 4.92FVFNTLSGNGNDD602 pKa = 4.27VITDD606 pKa = 3.8FQVGIDD612 pKa = 3.83HH613 pKa = 6.88IKK615 pKa = 10.83LVGLDD620 pKa = 3.69LGHH623 pKa = 6.97NPMQNLDD630 pKa = 3.42IRR632 pKa = 11.84DD633 pKa = 3.85TANGAVIEE641 pKa = 4.43YY642 pKa = 10.49GDD644 pKa = 3.66FTITVSGVSSHH655 pKa = 6.63SLTADD660 pKa = 3.26DD661 pKa = 6.7FIFF664 pKa = 4.65
Molecular weight: 67.42 kDa
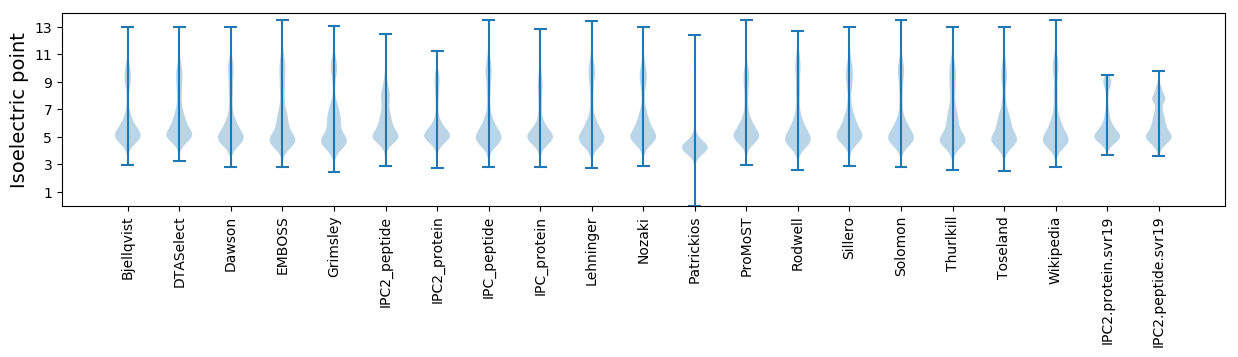
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I6IRU3|A0A6I6IRU3_9RHOB Ribosome-binding factor A OS=Roseovarius faecimaris OX=2494550 GN=rbfA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1185881 |
28 |
3335 |
317.3 |
34.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.115 ± 0.059 | 0.983 ± 0.012 |
6.029 ± 0.04 | 6.157 ± 0.039 |
3.703 ± 0.025 | 8.724 ± 0.05 |
2.166 ± 0.023 | 5.143 ± 0.028 |
3.154 ± 0.034 | 10.037 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.884 ± 0.021 | 2.586 ± 0.027 |
5.11 ± 0.027 | 3.222 ± 0.022 |
6.562 ± 0.045 | 5.11 ± 0.029 |
5.45 ± 0.042 | 7.081 ± 0.031 |
1.432 ± 0.016 | 2.353 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
