
Listeria phage P40
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
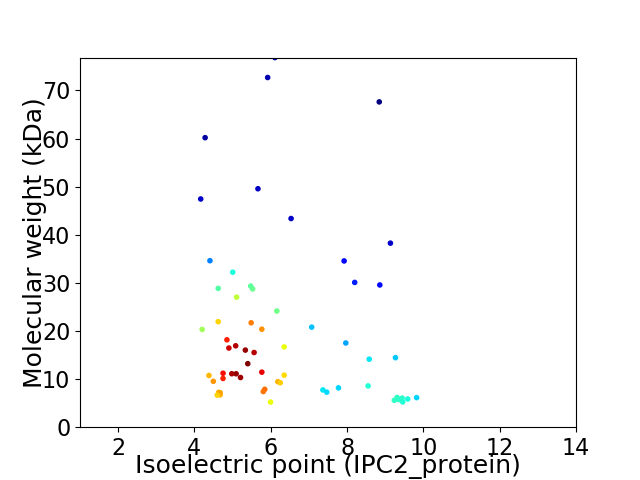
Virtual 2D-PAGE plot for 62 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6D7I9|B6D7I9_9CAUD Gp16 OS=Listeria phage P40 OX=560178 PE=4 SV=1
MM1 pKa = 7.46IPKK4 pKa = 10.3KK5 pKa = 9.82LTLINGRR12 pKa = 11.84GDD14 pKa = 3.54ILPLNDD20 pKa = 3.89NGSTGLFVLEE30 pKa = 4.11WSGLGVNINFDD41 pKa = 3.69LDD43 pKa = 3.55KK44 pKa = 10.8YY45 pKa = 10.29QASQIVKK52 pKa = 10.04NRR54 pKa = 11.84TLDD57 pKa = 3.34MQNFVVNIKK66 pKa = 10.3IGLNYY71 pKa = 10.39SATTPRR77 pKa = 11.84QIYY80 pKa = 10.23AGLIQFLNHH89 pKa = 6.02TPYY92 pKa = 10.55TLQSEE97 pKa = 4.56DD98 pKa = 3.84DD99 pKa = 3.6AGTYY103 pKa = 10.3LRR105 pKa = 11.84DD106 pKa = 3.6VQLQTMTLTDD116 pKa = 3.98LKK118 pKa = 10.29EE119 pKa = 4.0GQIFEE124 pKa = 4.36EE125 pKa = 4.35TLEE128 pKa = 4.64FICTSLWYY136 pKa = 10.73SEE138 pKa = 5.7DD139 pKa = 3.18IYY141 pKa = 10.97EE142 pKa = 4.22YY143 pKa = 10.36TPQDD147 pKa = 3.48LSIPADD153 pKa = 3.41TGKK156 pKa = 10.24VFDD159 pKa = 4.91KK160 pKa = 11.52NEE162 pKa = 3.92STTVKK167 pKa = 10.47NQGGYY172 pKa = 10.9GYY174 pKa = 10.86LLPTDD179 pKa = 4.27VNIAPYY185 pKa = 10.38EE186 pKa = 3.96PVGNGAITPDD196 pKa = 3.96MYY198 pKa = 11.26EE199 pKa = 4.47LGSPTITGAFLDD211 pKa = 3.69IDD213 pKa = 4.01EE214 pKa = 5.56LNVTFQAKK222 pKa = 10.05VYY224 pKa = 10.99LNGTLLFTGGTFDD237 pKa = 3.99NDD239 pKa = 3.23SGLFTINVPVTPVIGFNDD257 pKa = 3.59IVRR260 pKa = 11.84LDD262 pKa = 4.21LVTQISPDD270 pKa = 3.58TVLDD274 pKa = 3.95TNNVPLAQYY283 pKa = 10.19RR284 pKa = 11.84YY285 pKa = 9.08GYY287 pKa = 10.25IYY289 pKa = 10.62GITPNDD295 pKa = 3.08VKK297 pKa = 11.41GIFNVNNDD305 pKa = 3.74SVYY308 pKa = 10.44MGLTDD313 pKa = 4.95GSPCIITAYY322 pKa = 10.48DD323 pKa = 3.73PVSGITNPAVSVTVDD338 pKa = 3.33GNITQSDD345 pKa = 4.61RR346 pKa = 11.84YY347 pKa = 10.2VISIPNDD354 pKa = 3.39YY355 pKa = 10.25AWEE358 pKa = 4.11VSSIDD363 pKa = 3.1GTAYY367 pKa = 9.76CVRR370 pKa = 11.84YY371 pKa = 10.19NRR373 pKa = 11.84TTGVFEE379 pKa = 6.02NIYY382 pKa = 10.21QYY384 pKa = 11.29QDD386 pKa = 3.03QTKK389 pKa = 9.05TNFIKK394 pKa = 10.63IPVGDD399 pKa = 3.77SQVRR403 pKa = 11.84FGGINFDD410 pKa = 4.1NPGSYY415 pKa = 10.9AKK417 pKa = 10.22IIVRR421 pKa = 11.84RR422 pKa = 11.84EE423 pKa = 3.62RR424 pKa = 11.84LSIGG428 pKa = 3.23
MM1 pKa = 7.46IPKK4 pKa = 10.3KK5 pKa = 9.82LTLINGRR12 pKa = 11.84GDD14 pKa = 3.54ILPLNDD20 pKa = 3.89NGSTGLFVLEE30 pKa = 4.11WSGLGVNINFDD41 pKa = 3.69LDD43 pKa = 3.55KK44 pKa = 10.8YY45 pKa = 10.29QASQIVKK52 pKa = 10.04NRR54 pKa = 11.84TLDD57 pKa = 3.34MQNFVVNIKK66 pKa = 10.3IGLNYY71 pKa = 10.39SATTPRR77 pKa = 11.84QIYY80 pKa = 10.23AGLIQFLNHH89 pKa = 6.02TPYY92 pKa = 10.55TLQSEE97 pKa = 4.56DD98 pKa = 3.84DD99 pKa = 3.6AGTYY103 pKa = 10.3LRR105 pKa = 11.84DD106 pKa = 3.6VQLQTMTLTDD116 pKa = 3.98LKK118 pKa = 10.29EE119 pKa = 4.0GQIFEE124 pKa = 4.36EE125 pKa = 4.35TLEE128 pKa = 4.64FICTSLWYY136 pKa = 10.73SEE138 pKa = 5.7DD139 pKa = 3.18IYY141 pKa = 10.97EE142 pKa = 4.22YY143 pKa = 10.36TPQDD147 pKa = 3.48LSIPADD153 pKa = 3.41TGKK156 pKa = 10.24VFDD159 pKa = 4.91KK160 pKa = 11.52NEE162 pKa = 3.92STTVKK167 pKa = 10.47NQGGYY172 pKa = 10.9GYY174 pKa = 10.86LLPTDD179 pKa = 4.27VNIAPYY185 pKa = 10.38EE186 pKa = 3.96PVGNGAITPDD196 pKa = 3.96MYY198 pKa = 11.26EE199 pKa = 4.47LGSPTITGAFLDD211 pKa = 3.69IDD213 pKa = 4.01EE214 pKa = 5.56LNVTFQAKK222 pKa = 10.05VYY224 pKa = 10.99LNGTLLFTGGTFDD237 pKa = 3.99NDD239 pKa = 3.23SGLFTINVPVTPVIGFNDD257 pKa = 3.59IVRR260 pKa = 11.84LDD262 pKa = 4.21LVTQISPDD270 pKa = 3.58TVLDD274 pKa = 3.95TNNVPLAQYY283 pKa = 10.19RR284 pKa = 11.84YY285 pKa = 9.08GYY287 pKa = 10.25IYY289 pKa = 10.62GITPNDD295 pKa = 3.08VKK297 pKa = 11.41GIFNVNNDD305 pKa = 3.74SVYY308 pKa = 10.44MGLTDD313 pKa = 4.95GSPCIITAYY322 pKa = 10.48DD323 pKa = 3.73PVSGITNPAVSVTVDD338 pKa = 3.33GNITQSDD345 pKa = 4.61RR346 pKa = 11.84YY347 pKa = 10.2VISIPNDD354 pKa = 3.39YY355 pKa = 10.25AWEE358 pKa = 4.11VSSIDD363 pKa = 3.1GTAYY367 pKa = 9.76CVRR370 pKa = 11.84YY371 pKa = 10.19NRR373 pKa = 11.84TTGVFEE379 pKa = 6.02NIYY382 pKa = 10.21QYY384 pKa = 11.29QDD386 pKa = 3.03QTKK389 pKa = 9.05TNFIKK394 pKa = 10.63IPVGDD399 pKa = 3.77SQVRR403 pKa = 11.84FGGINFDD410 pKa = 4.1NPGSYY415 pKa = 10.9AKK417 pKa = 10.22IIVRR421 pKa = 11.84RR422 pKa = 11.84EE423 pKa = 3.62RR424 pKa = 11.84LSIGG428 pKa = 3.23
Molecular weight: 47.44 kDa
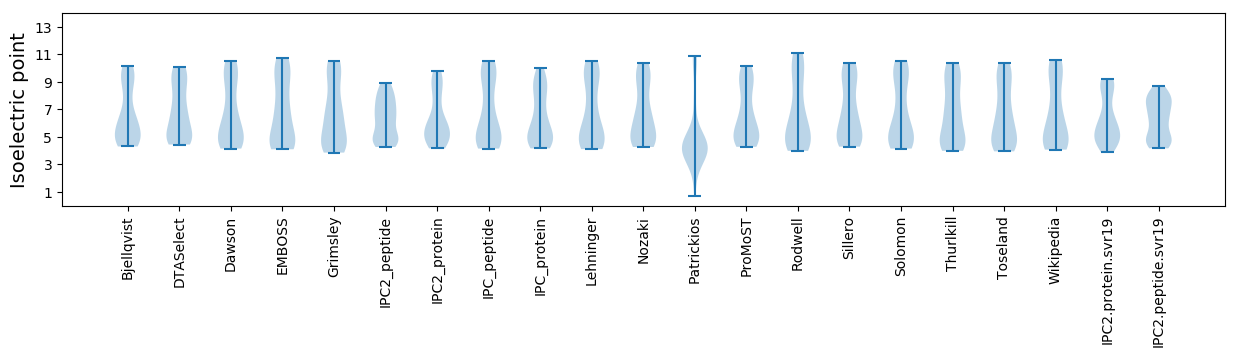
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6D7J8|B6D7J8_9CAUD Gp25 OS=Listeria phage P40 OX=560178 PE=4 SV=1
MM1 pKa = 7.52VVILTFFIVGMCVGMFNVYY20 pKa = 10.18HH21 pKa = 5.63YY22 pKa = 10.03VAKK25 pKa = 9.95PALQEE30 pKa = 3.76LRR32 pKa = 11.84LLKK35 pKa = 10.78GEE37 pKa = 4.51PLPKK41 pKa = 9.47RR42 pKa = 11.84AKK44 pKa = 10.23FSIKK48 pKa = 8.74ITRR51 pKa = 3.45
MM1 pKa = 7.52VVILTFFIVGMCVGMFNVYY20 pKa = 10.18HH21 pKa = 5.63YY22 pKa = 10.03VAKK25 pKa = 9.95PALQEE30 pKa = 3.76LRR32 pKa = 11.84LLKK35 pKa = 10.78GEE37 pKa = 4.51PLPKK41 pKa = 9.47RR42 pKa = 11.84AKK44 pKa = 10.23FSIKK48 pKa = 8.74ITRR51 pKa = 3.45
Molecular weight: 5.87 kDa
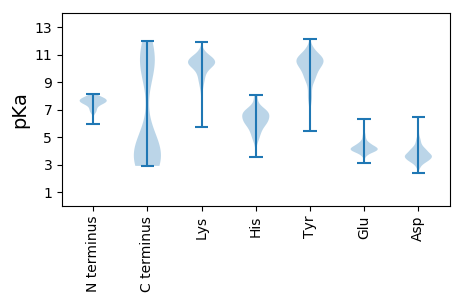
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11025 |
43 |
668 |
177.8 |
20.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.56 ± 0.353 | 0.744 ± 0.087 |
6.567 ± 0.316 | 7.138 ± 0.464 |
4.218 ± 0.248 | 6.113 ± 0.447 |
1.723 ± 0.177 | 7.356 ± 0.235 |
8.19 ± 0.471 | 8.444 ± 0.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.821 ± 0.27 | 5.905 ± 0.293 |
3.338 ± 0.213 | 3.42 ± 0.192 |
4.127 ± 0.274 | 5.651 ± 0.214 |
6.24 ± 0.329 | 6.948 ± 0.262 |
1.07 ± 0.15 | 4.426 ± 0.244 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
