
Halomonas sp. JB380
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; unclassified Halomonas
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
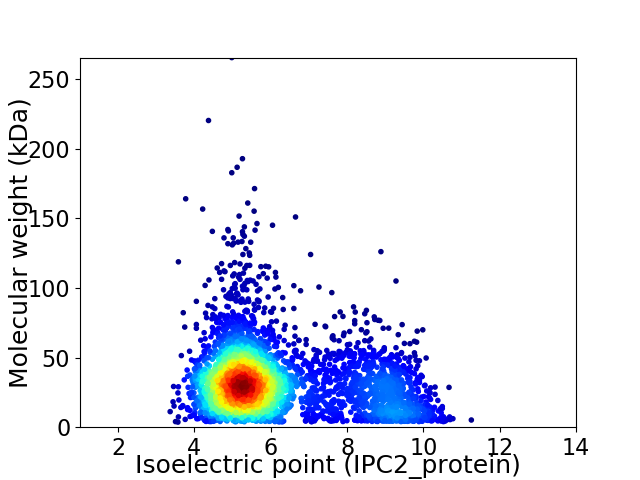
Virtual 2D-PAGE plot for 3787 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
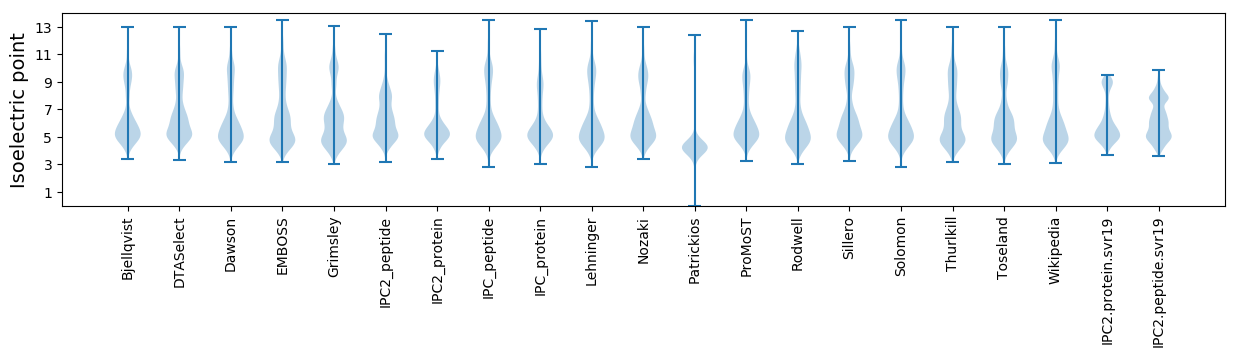
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R4HZX8|A0A1R4HZX8_9GAMM Uncharacterized protein OS=Halomonas sp. JB380 OX=1434831 GN=CZ787_09525 PE=3 SV=1
MM1 pKa = 7.49AKK3 pKa = 9.96SSPFALIVVAVGGAVVGFLAAQTLSSDD30 pKa = 3.61ASSSSSVASQSAEE43 pKa = 3.8QTPTADD49 pKa = 3.57ALVGLDD55 pKa = 4.3SITLGEE61 pKa = 4.17RR62 pKa = 11.84LRR64 pKa = 11.84GEE66 pKa = 3.89ITASDD71 pKa = 4.4LLNGNDD77 pKa = 3.28GSRR80 pKa = 11.84YY81 pKa = 9.31DD82 pKa = 3.87RR83 pKa = 11.84YY84 pKa = 11.01MMTLDD89 pKa = 3.73EE90 pKa = 4.77SDD92 pKa = 4.93LIEE95 pKa = 4.41FSLRR99 pKa = 11.84GALNGIVALYY109 pKa = 10.48DD110 pKa = 3.74DD111 pKa = 4.4QLQLLSVAPTARR123 pKa = 11.84HH124 pKa = 6.21RR125 pKa = 11.84IEE127 pKa = 3.72QAGDD131 pKa = 3.35YY132 pKa = 9.16MVVVSGADD140 pKa = 3.16SSSYY144 pKa = 11.18GPYY147 pKa = 8.98TLNSRR152 pKa = 11.84VVEE155 pKa = 4.22LSDD158 pKa = 3.86STVLTLDD165 pKa = 3.91APLDD169 pKa = 3.5SWLDD173 pKa = 3.8GTSSEE178 pKa = 4.26YY179 pKa = 10.76TLTIEE184 pKa = 4.32EE185 pKa = 4.13AGMYY189 pKa = 8.38QLEE192 pKa = 4.23MRR194 pKa = 11.84SDD196 pKa = 3.37EE197 pKa = 3.9FDD199 pKa = 3.64AYY201 pKa = 11.37LEE203 pKa = 3.99LSGPNGYY210 pKa = 9.4FRR212 pKa = 11.84EE213 pKa = 4.71DD214 pKa = 3.79DD215 pKa = 3.99DD216 pKa = 5.1SAGNLDD222 pKa = 3.72ARR224 pKa = 11.84ISDD227 pKa = 3.73FLVPGDD233 pKa = 3.79YY234 pKa = 10.38TVTARR239 pKa = 11.84TSYY242 pKa = 11.52NEE244 pKa = 3.89GAGLFTLTAEE254 pKa = 4.29SRR256 pKa = 11.84EE257 pKa = 4.05LPGDD261 pKa = 3.64GEE263 pKa = 4.27LRR265 pKa = 11.84NDD267 pKa = 3.69GSIVPNDD274 pKa = 4.03SLNGLYY280 pKa = 10.33SGQNLSYY287 pKa = 10.3QVEE290 pKa = 4.08IEE292 pKa = 4.16EE293 pKa = 4.28AGMYY297 pKa = 9.55QIDD300 pKa = 4.38MMSDD304 pKa = 3.94DD305 pKa = 3.3IDD307 pKa = 4.9AYY309 pKa = 11.12LVLEE313 pKa = 5.06GPDD316 pKa = 3.27GYY318 pKa = 11.41YY319 pKa = 9.95RR320 pKa = 11.84EE321 pKa = 4.48NDD323 pKa = 3.59DD324 pKa = 5.81GGDD327 pKa = 3.62NLNARR332 pKa = 11.84IADD335 pKa = 3.78FFEE338 pKa = 5.12PGTYY342 pKa = 10.22QLTARR347 pKa = 11.84TAYY350 pKa = 10.31GSDD353 pKa = 2.91SGLFRR358 pKa = 11.84ITLEE362 pKa = 4.08PQEE365 pKa = 4.38LPEE368 pKa = 4.34GVEE371 pKa = 4.13LRR373 pKa = 11.84NSGAITLGEE382 pKa = 4.39TLNGWYY388 pKa = 8.94TGEE391 pKa = 3.99SLVYY395 pKa = 9.56EE396 pKa = 4.37LEE398 pKa = 4.16LAEE401 pKa = 4.63SSLVTINMRR410 pKa = 11.84SSDD413 pKa = 3.41FDD415 pKa = 4.08AYY417 pKa = 11.3LEE419 pKa = 4.25LEE421 pKa = 4.46GEE423 pKa = 4.7GVAVSDD429 pKa = 4.51DD430 pKa = 3.9DD431 pKa = 5.13GGAGTDD437 pKa = 3.6AQLQQQLLPGTYY449 pKa = 8.77TVIARR454 pKa = 11.84GFSTASSGLFEE465 pKa = 6.53LDD467 pKa = 3.16VTAEE471 pKa = 4.07PAQMQPNMM479 pKa = 4.11
MM1 pKa = 7.49AKK3 pKa = 9.96SSPFALIVVAVGGAVVGFLAAQTLSSDD30 pKa = 3.61ASSSSSVASQSAEE43 pKa = 3.8QTPTADD49 pKa = 3.57ALVGLDD55 pKa = 4.3SITLGEE61 pKa = 4.17RR62 pKa = 11.84LRR64 pKa = 11.84GEE66 pKa = 3.89ITASDD71 pKa = 4.4LLNGNDD77 pKa = 3.28GSRR80 pKa = 11.84YY81 pKa = 9.31DD82 pKa = 3.87RR83 pKa = 11.84YY84 pKa = 11.01MMTLDD89 pKa = 3.73EE90 pKa = 4.77SDD92 pKa = 4.93LIEE95 pKa = 4.41FSLRR99 pKa = 11.84GALNGIVALYY109 pKa = 10.48DD110 pKa = 3.74DD111 pKa = 4.4QLQLLSVAPTARR123 pKa = 11.84HH124 pKa = 6.21RR125 pKa = 11.84IEE127 pKa = 3.72QAGDD131 pKa = 3.35YY132 pKa = 9.16MVVVSGADD140 pKa = 3.16SSSYY144 pKa = 11.18GPYY147 pKa = 8.98TLNSRR152 pKa = 11.84VVEE155 pKa = 4.22LSDD158 pKa = 3.86STVLTLDD165 pKa = 3.91APLDD169 pKa = 3.5SWLDD173 pKa = 3.8GTSSEE178 pKa = 4.26YY179 pKa = 10.76TLTIEE184 pKa = 4.32EE185 pKa = 4.13AGMYY189 pKa = 8.38QLEE192 pKa = 4.23MRR194 pKa = 11.84SDD196 pKa = 3.37EE197 pKa = 3.9FDD199 pKa = 3.64AYY201 pKa = 11.37LEE203 pKa = 3.99LSGPNGYY210 pKa = 9.4FRR212 pKa = 11.84EE213 pKa = 4.71DD214 pKa = 3.79DD215 pKa = 3.99DD216 pKa = 5.1SAGNLDD222 pKa = 3.72ARR224 pKa = 11.84ISDD227 pKa = 3.73FLVPGDD233 pKa = 3.79YY234 pKa = 10.38TVTARR239 pKa = 11.84TSYY242 pKa = 11.52NEE244 pKa = 3.89GAGLFTLTAEE254 pKa = 4.29SRR256 pKa = 11.84EE257 pKa = 4.05LPGDD261 pKa = 3.64GEE263 pKa = 4.27LRR265 pKa = 11.84NDD267 pKa = 3.69GSIVPNDD274 pKa = 4.03SLNGLYY280 pKa = 10.33SGQNLSYY287 pKa = 10.3QVEE290 pKa = 4.08IEE292 pKa = 4.16EE293 pKa = 4.28AGMYY297 pKa = 9.55QIDD300 pKa = 4.38MMSDD304 pKa = 3.94DD305 pKa = 3.3IDD307 pKa = 4.9AYY309 pKa = 11.12LVLEE313 pKa = 5.06GPDD316 pKa = 3.27GYY318 pKa = 11.41YY319 pKa = 9.95RR320 pKa = 11.84EE321 pKa = 4.48NDD323 pKa = 3.59DD324 pKa = 5.81GGDD327 pKa = 3.62NLNARR332 pKa = 11.84IADD335 pKa = 3.78FFEE338 pKa = 5.12PGTYY342 pKa = 10.22QLTARR347 pKa = 11.84TAYY350 pKa = 10.31GSDD353 pKa = 2.91SGLFRR358 pKa = 11.84ITLEE362 pKa = 4.08PQEE365 pKa = 4.38LPEE368 pKa = 4.34GVEE371 pKa = 4.13LRR373 pKa = 11.84NSGAITLGEE382 pKa = 4.39TLNGWYY388 pKa = 8.94TGEE391 pKa = 3.99SLVYY395 pKa = 9.56EE396 pKa = 4.37LEE398 pKa = 4.16LAEE401 pKa = 4.63SSLVTINMRR410 pKa = 11.84SSDD413 pKa = 3.41FDD415 pKa = 4.08AYY417 pKa = 11.3LEE419 pKa = 4.25LEE421 pKa = 4.46GEE423 pKa = 4.7GVAVSDD429 pKa = 4.51DD430 pKa = 3.9DD431 pKa = 5.13GGAGTDD437 pKa = 3.6AQLQQQLLPGTYY449 pKa = 8.77TVIARR454 pKa = 11.84GFSTASSGLFEE465 pKa = 6.53LDD467 pKa = 3.16VTAEE471 pKa = 4.07PAQMQPNMM479 pKa = 4.11
Molecular weight: 51.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R4HUY1|A0A1R4HUY1_9GAMM ATP synthase subunit delta OS=Halomonas sp. JB380 OX=1434831 GN=atpH PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.21NGRR28 pKa = 11.84AVISRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.97GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.21NGRR28 pKa = 11.84AVISRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.97GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1169929 |
37 |
2452 |
308.9 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.754 ± 0.041 | 0.924 ± 0.014 |
5.381 ± 0.037 | 6.176 ± 0.039 |
3.61 ± 0.027 | 7.708 ± 0.04 |
2.422 ± 0.021 | 5.284 ± 0.032 |
3.198 ± 0.036 | 11.248 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.651 ± 0.019 | 3.152 ± 0.025 |
4.707 ± 0.025 | 4.346 ± 0.037 |
6.142 ± 0.037 | 5.911 ± 0.027 |
5.287 ± 0.025 | 7.151 ± 0.034 |
1.424 ± 0.018 | 2.524 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
