
Flavobacteriaceae bacterium CWB-1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Paucihalobacter
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
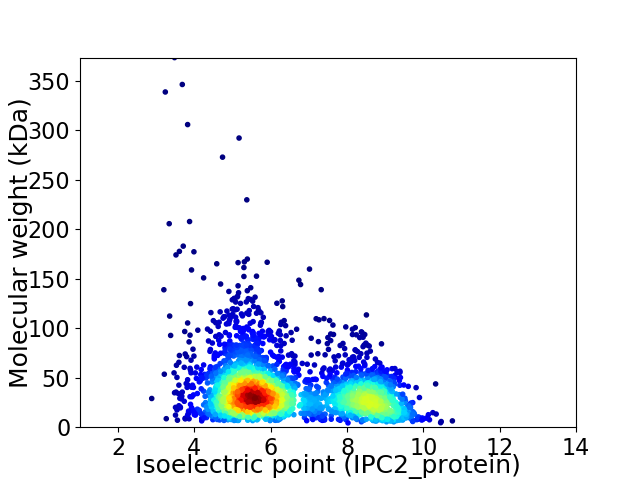
Virtual 2D-PAGE plot for 3060 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A506PDI3|A0A506PDI3_9FLAO Response regulator transcription factor OS=Flavobacteriaceae bacterium CWB-1 OX=2567861 GN=FJ651_14300 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.17KK3 pKa = 10.63LSILGLFIFALISFNSCEE21 pKa = 4.27TEE23 pKa = 3.86DD24 pKa = 3.98DD25 pKa = 4.04VVFIANQADD34 pKa = 3.87FAFTNSFSAQYY45 pKa = 11.08VLTPIAAQNLGEE57 pKa = 4.16RR58 pKa = 11.84FTWNTPDD65 pKa = 3.23VGVPTNITYY74 pKa = 10.39QLQRR78 pKa = 11.84SIAGDD83 pKa = 3.4FSDD86 pKa = 4.33AVTVGATSSNEE97 pKa = 3.43IAITIGDD104 pKa = 4.11MLVIAAQAGLDD115 pKa = 3.54SDD117 pKa = 5.04PNTEE121 pKa = 4.21NPNTGQVSFRR131 pKa = 11.84VRR133 pKa = 11.84STVGDD138 pKa = 3.84GGLEE142 pKa = 4.33TYY144 pKa = 11.25SNTAVLTLVLPEE156 pKa = 4.56DD157 pKa = 3.97TTTAPAVCDD166 pKa = 3.58HH167 pKa = 7.07DD168 pKa = 4.16QLFLVGAGVKK178 pKa = 9.63FAGWSWDD185 pKa = 3.4TPQVINCSGNGVYY198 pKa = 9.58TGNVAFTNNVDD209 pKa = 3.04GDD211 pKa = 4.09GNFRR215 pKa = 11.84FFSTATDD222 pKa = 3.19WGSGTNFPGYY232 pKa = 10.94ANDD235 pKa = 4.95GYY237 pKa = 9.6TIDD240 pKa = 5.1SRR242 pKa = 11.84FEE244 pKa = 3.88DD245 pKa = 4.66AQDD248 pKa = 3.41GDD250 pKa = 4.06NNFLFTGASGLYY262 pKa = 10.0FLTIDD267 pKa = 5.48DD268 pKa = 3.9INKK271 pKa = 9.25TITLDD276 pKa = 3.78DD277 pKa = 3.94PTPTGSCDD285 pKa = 5.0LDD287 pKa = 3.6QLWAVGAGLPDD298 pKa = 5.13AGWSWDD304 pKa = 3.52TPIQFMCEE312 pKa = 3.98GNGVYY317 pKa = 9.97QGWVNLSPEE326 pKa = 3.71NDD328 pKa = 3.17GNFRR332 pKa = 11.84FFNTATDD339 pKa = 3.36WGSGTNYY346 pKa = 9.96PGFISQGYY354 pKa = 7.77TIDD357 pKa = 4.92NRR359 pKa = 11.84FQDD362 pKa = 4.58AQDD365 pKa = 3.56GDD367 pKa = 4.12NNFQFIGTAGQYY379 pKa = 9.36FLKK382 pKa = 10.13IDD384 pKa = 4.63TINKK388 pKa = 8.27TITLEE393 pKa = 3.96
MM1 pKa = 7.61KK2 pKa = 10.17KK3 pKa = 10.63LSILGLFIFALISFNSCEE21 pKa = 4.27TEE23 pKa = 3.86DD24 pKa = 3.98DD25 pKa = 4.04VVFIANQADD34 pKa = 3.87FAFTNSFSAQYY45 pKa = 11.08VLTPIAAQNLGEE57 pKa = 4.16RR58 pKa = 11.84FTWNTPDD65 pKa = 3.23VGVPTNITYY74 pKa = 10.39QLQRR78 pKa = 11.84SIAGDD83 pKa = 3.4FSDD86 pKa = 4.33AVTVGATSSNEE97 pKa = 3.43IAITIGDD104 pKa = 4.11MLVIAAQAGLDD115 pKa = 3.54SDD117 pKa = 5.04PNTEE121 pKa = 4.21NPNTGQVSFRR131 pKa = 11.84VRR133 pKa = 11.84STVGDD138 pKa = 3.84GGLEE142 pKa = 4.33TYY144 pKa = 11.25SNTAVLTLVLPEE156 pKa = 4.56DD157 pKa = 3.97TTTAPAVCDD166 pKa = 3.58HH167 pKa = 7.07DD168 pKa = 4.16QLFLVGAGVKK178 pKa = 9.63FAGWSWDD185 pKa = 3.4TPQVINCSGNGVYY198 pKa = 9.58TGNVAFTNNVDD209 pKa = 3.04GDD211 pKa = 4.09GNFRR215 pKa = 11.84FFSTATDD222 pKa = 3.19WGSGTNFPGYY232 pKa = 10.94ANDD235 pKa = 4.95GYY237 pKa = 9.6TIDD240 pKa = 5.1SRR242 pKa = 11.84FEE244 pKa = 3.88DD245 pKa = 4.66AQDD248 pKa = 3.41GDD250 pKa = 4.06NNFLFTGASGLYY262 pKa = 10.0FLTIDD267 pKa = 5.48DD268 pKa = 3.9INKK271 pKa = 9.25TITLDD276 pKa = 3.78DD277 pKa = 3.94PTPTGSCDD285 pKa = 5.0LDD287 pKa = 3.6QLWAVGAGLPDD298 pKa = 5.13AGWSWDD304 pKa = 3.52TPIQFMCEE312 pKa = 3.98GNGVYY317 pKa = 9.97QGWVNLSPEE326 pKa = 3.71NDD328 pKa = 3.17GNFRR332 pKa = 11.84FFNTATDD339 pKa = 3.36WGSGTNYY346 pKa = 9.96PGFISQGYY354 pKa = 7.77TIDD357 pKa = 4.92NRR359 pKa = 11.84FQDD362 pKa = 4.58AQDD365 pKa = 3.56GDD367 pKa = 4.12NNFQFIGTAGQYY379 pKa = 9.36FLKK382 pKa = 10.13IDD384 pKa = 4.63TINKK388 pKa = 8.27TITLEE393 pKa = 3.96
Molecular weight: 42.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A506PLT4|A0A506PLT4_9FLAO Pyruvate kinase OS=Flavobacteriaceae bacterium CWB-1 OX=2567861 GN=pyk PE=3 SV=1
MM1 pKa = 7.45TFPTVKK7 pKa = 10.0NVKK10 pKa = 9.08VAGNRR15 pKa = 11.84RR16 pKa = 11.84SAQWPVRR23 pKa = 11.84VLFEE27 pKa = 4.9HH28 pKa = 6.05STSHH32 pKa = 6.9QLLCYY37 pKa = 10.03LDD39 pKa = 4.53SFEE42 pKa = 5.33LRR44 pKa = 11.84NPHH47 pKa = 5.89RR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 9.41AAKK53 pKa = 9.58RR54 pKa = 11.84YY55 pKa = 9.74HH56 pKa = 6.26PFEE59 pKa = 5.3KK60 pKa = 10.72NILII64 pKa = 4.45
MM1 pKa = 7.45TFPTVKK7 pKa = 10.0NVKK10 pKa = 9.08VAGNRR15 pKa = 11.84RR16 pKa = 11.84SAQWPVRR23 pKa = 11.84VLFEE27 pKa = 4.9HH28 pKa = 6.05STSHH32 pKa = 6.9QLLCYY37 pKa = 10.03LDD39 pKa = 4.53SFEE42 pKa = 5.33LRR44 pKa = 11.84NPHH47 pKa = 5.89RR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 9.41AAKK53 pKa = 9.58RR54 pKa = 11.84YY55 pKa = 9.74HH56 pKa = 6.26PFEE59 pKa = 5.3KK60 pKa = 10.72NILII64 pKa = 4.45
Molecular weight: 7.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1058090 |
38 |
3586 |
345.8 |
39.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.441 ± 0.047 | 0.751 ± 0.018 |
5.493 ± 0.034 | 6.276 ± 0.047 |
5.387 ± 0.039 | 6.282 ± 0.05 |
1.827 ± 0.025 | 8.043 ± 0.041 |
7.123 ± 0.072 | 9.474 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.026 | 6.684 ± 0.067 |
3.461 ± 0.026 | 3.679 ± 0.025 |
3.388 ± 0.031 | 6.335 ± 0.036 |
5.874 ± 0.07 | 6.279 ± 0.037 |
1.063 ± 0.016 | 3.998 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
