
Terrimonas sp. NS-102
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Terrimonas; unclassified Terrimonas
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
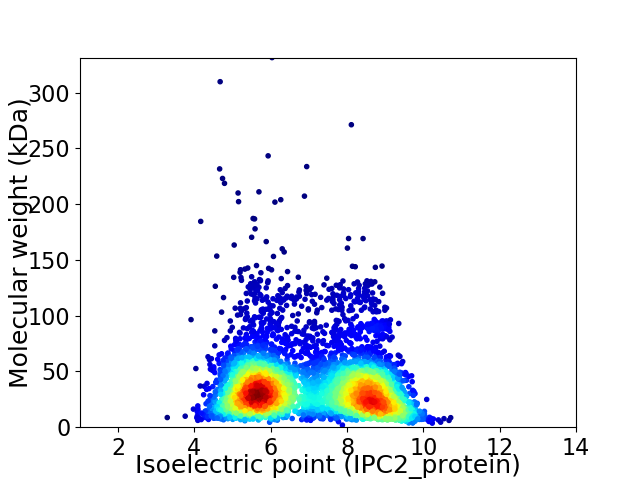
Virtual 2D-PAGE plot for 5019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T7Q2E9|A0A2T7Q2E9_9BACT IS66 family transposase OS=Terrimonas sp. NS-102 OX=2171976 GN=DC498_24570 PE=4 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84YY3 pKa = 9.61IIFTILLFVQLGVYY17 pKa = 9.17AQSTGDD23 pKa = 3.63YY24 pKa = 10.08RR25 pKa = 11.84SKK27 pKa = 11.03QSGNWEE33 pKa = 4.06DD34 pKa = 3.29AGSWEE39 pKa = 4.63TYY41 pKa = 10.85NGTSWVAATNYY52 pKa = 6.7PTPSDD57 pKa = 3.39GTITIRR63 pKa = 11.84SPHH66 pKa = 7.0ILTQGSSFTINDD78 pKa = 3.06VTVEE82 pKa = 4.04VGATLNLNDD91 pKa = 4.25GNVNSSGSSTADD103 pKa = 2.98LRR105 pKa = 11.84VYY107 pKa = 9.73GTVNHH112 pKa = 6.51NANSQGGCPIFEE124 pKa = 4.43IYY126 pKa = 10.56NGGVYY131 pKa = 9.86NWNGGNYY138 pKa = 9.88ACNTIKK144 pKa = 10.42ILSGGTMNFNVSGNPYY160 pKa = 10.46LNEE163 pKa = 4.01TNITNDD169 pKa = 2.85GVINFNSGGFYY180 pKa = 10.58AAINTVWGNLVNNAGGVINKK200 pKa = 9.61NNDD203 pKa = 3.37NIFFASGSPFNFIQNGSLNINAGRR227 pKa = 11.84LHH229 pKa = 7.04IDD231 pKa = 3.13YY232 pKa = 10.96LNFSNTGQLSIANNAEE248 pKa = 4.5LVCSGTPLMLSGTLNVIEE266 pKa = 5.0KK267 pKa = 10.04VSPSNGSNVIISGNFSGNFSTVNLPIGYY295 pKa = 9.34SITVNPSDD303 pKa = 5.13VILNYY308 pKa = 10.39NDD310 pKa = 5.48DD311 pKa = 3.79MDD313 pKa = 6.87DD314 pKa = 4.84DD315 pKa = 4.52GVKK318 pKa = 10.76NKK320 pKa = 10.21DD321 pKa = 3.06DD322 pKa = 4.63CAPKK326 pKa = 10.61DD327 pKa = 3.64PNKK330 pKa = 9.35WRR332 pKa = 11.84SAEE335 pKa = 4.02FYY337 pKa = 10.18IDD339 pKa = 4.17KK340 pKa = 11.01DD341 pKa = 3.26SDD343 pKa = 3.91GYY345 pKa = 11.37DD346 pKa = 3.44GGKK349 pKa = 8.24HH350 pKa = 4.2TVCYY354 pKa = 9.37GQNIPSGYY362 pKa = 9.46IQTTKK367 pKa = 10.87GSDD370 pKa = 3.87CNDD373 pKa = 2.85NDD375 pKa = 4.55ANINPTTVWYY385 pKa = 10.62KK386 pKa = 10.87DD387 pKa = 3.35ADD389 pKa = 3.62NDD391 pKa = 4.49GYY393 pKa = 11.71SDD395 pKa = 3.8GTTKK399 pKa = 8.92TQCDD403 pKa = 3.31QPAGYY408 pKa = 9.59KK409 pKa = 10.36LKK411 pKa = 10.9AQLTATNGDD420 pKa = 4.3CKK422 pKa = 11.18DD423 pKa = 4.15DD424 pKa = 4.65DD425 pKa = 4.08ATIHH429 pKa = 6.62PGAPEE434 pKa = 3.61ICGNGIDD441 pKa = 4.59EE442 pKa = 5.21DD443 pKa = 4.77CDD445 pKa = 4.1SKK447 pKa = 11.74DD448 pKa = 3.62AVCVPTDD455 pKa = 3.29SDD457 pKa = 4.03GDD459 pKa = 4.09GVSDD463 pKa = 6.01NEE465 pKa = 4.44DD466 pKa = 3.67CSPNDD471 pKa = 3.21NKK473 pKa = 10.15VWRR476 pKa = 11.84TVTLYY481 pKa = 11.25ADD483 pKa = 4.38FDD485 pKa = 5.08SDD487 pKa = 3.78GKK489 pKa = 9.87PVAFGSEE496 pKa = 4.09VCIGADD502 pKa = 2.94IPQGYY507 pKa = 9.11SEE509 pKa = 5.4SPGSDD514 pKa = 3.51CDD516 pKa = 5.54DD517 pKa = 3.82NDD519 pKa = 3.41NTVWRR524 pKa = 11.84TAILYY529 pKa = 9.88IDD531 pKa = 3.97SDD533 pKa = 4.05RR534 pKa = 11.84DD535 pKa = 3.85GEE537 pKa = 4.68SVGAGIEE544 pKa = 4.0KK545 pKa = 10.57CIGNDD550 pKa = 2.72IPFGYY555 pKa = 8.87TEE557 pKa = 5.11SPGSDD562 pKa = 3.48CNDD565 pKa = 3.49NNPDD569 pKa = 3.4IYY571 pKa = 10.92HH572 pKa = 7.0GATEE576 pKa = 4.12ICDD579 pKa = 3.84GVDD582 pKa = 3.45NNCDD586 pKa = 3.4GQIDD590 pKa = 4.0EE591 pKa = 5.99GLLFWIYY598 pKa = 10.46PDD600 pKa = 4.36GDD602 pKa = 3.45GDD604 pKa = 4.53GYY606 pKa = 10.2GTEE609 pKa = 3.84EE610 pKa = 4.24GKK612 pKa = 10.29IYY614 pKa = 10.48SCNAPYY620 pKa = 10.55GYY622 pKa = 10.84ADD624 pKa = 4.5RR625 pKa = 11.84NGDD628 pKa = 4.75CKK630 pKa = 11.31DD631 pKa = 4.04DD632 pKa = 5.09DD633 pKa = 3.84NTINPGVEE641 pKa = 4.44EE642 pKa = 4.3ICDD645 pKa = 3.73DD646 pKa = 5.88GIDD649 pKa = 3.79NDD651 pKa = 4.45CDD653 pKa = 4.12GEE655 pKa = 4.05IDD657 pKa = 3.74EE658 pKa = 5.22GCSVSEE664 pKa = 3.8PTEE667 pKa = 4.71FYY669 pKa = 11.1SKK671 pKa = 9.55PTGDD675 pKa = 3.76LHH677 pKa = 7.35NVATWGVNPDD687 pKa = 3.35GSGTQPADD695 pKa = 3.86FGAGKK700 pKa = 8.29TFNLANRR707 pKa = 11.84AGNYY711 pKa = 8.47TMTGNWTVLGTLVNSSGSQLKK732 pKa = 10.43INGYY736 pKa = 6.21TLSLTTLTGAGTLTGSTTSSLIITGTGGGNFGNINFTSGGGMLKK780 pKa = 10.6AFTLNRR786 pKa = 11.84SGTGAAATIGTALAVYY802 pKa = 9.99DD803 pKa = 4.18VLTITSGALTTGGKK817 pKa = 8.08LTLKK821 pKa = 9.52STATNTARR829 pKa = 11.84VAPVTGTISGNVTVEE844 pKa = 3.79RR845 pKa = 11.84YY846 pKa = 9.59IPARR850 pKa = 11.84RR851 pKa = 11.84AWRR854 pKa = 11.84LMNAPVGGTQTINQAWQEE872 pKa = 4.27GVTTASPNPNPAPGYY887 pKa = 7.66GTYY890 pKa = 8.98VTVGSVANGFDD901 pKa = 4.01QNILGQSTSSLKK913 pKa = 10.96SFF915 pKa = 4.16
MM1 pKa = 7.96RR2 pKa = 11.84YY3 pKa = 9.61IIFTILLFVQLGVYY17 pKa = 9.17AQSTGDD23 pKa = 3.63YY24 pKa = 10.08RR25 pKa = 11.84SKK27 pKa = 11.03QSGNWEE33 pKa = 4.06DD34 pKa = 3.29AGSWEE39 pKa = 4.63TYY41 pKa = 10.85NGTSWVAATNYY52 pKa = 6.7PTPSDD57 pKa = 3.39GTITIRR63 pKa = 11.84SPHH66 pKa = 7.0ILTQGSSFTINDD78 pKa = 3.06VTVEE82 pKa = 4.04VGATLNLNDD91 pKa = 4.25GNVNSSGSSTADD103 pKa = 2.98LRR105 pKa = 11.84VYY107 pKa = 9.73GTVNHH112 pKa = 6.51NANSQGGCPIFEE124 pKa = 4.43IYY126 pKa = 10.56NGGVYY131 pKa = 9.86NWNGGNYY138 pKa = 9.88ACNTIKK144 pKa = 10.42ILSGGTMNFNVSGNPYY160 pKa = 10.46LNEE163 pKa = 4.01TNITNDD169 pKa = 2.85GVINFNSGGFYY180 pKa = 10.58AAINTVWGNLVNNAGGVINKK200 pKa = 9.61NNDD203 pKa = 3.37NIFFASGSPFNFIQNGSLNINAGRR227 pKa = 11.84LHH229 pKa = 7.04IDD231 pKa = 3.13YY232 pKa = 10.96LNFSNTGQLSIANNAEE248 pKa = 4.5LVCSGTPLMLSGTLNVIEE266 pKa = 5.0KK267 pKa = 10.04VSPSNGSNVIISGNFSGNFSTVNLPIGYY295 pKa = 9.34SITVNPSDD303 pKa = 5.13VILNYY308 pKa = 10.39NDD310 pKa = 5.48DD311 pKa = 3.79MDD313 pKa = 6.87DD314 pKa = 4.84DD315 pKa = 4.52GVKK318 pKa = 10.76NKK320 pKa = 10.21DD321 pKa = 3.06DD322 pKa = 4.63CAPKK326 pKa = 10.61DD327 pKa = 3.64PNKK330 pKa = 9.35WRR332 pKa = 11.84SAEE335 pKa = 4.02FYY337 pKa = 10.18IDD339 pKa = 4.17KK340 pKa = 11.01DD341 pKa = 3.26SDD343 pKa = 3.91GYY345 pKa = 11.37DD346 pKa = 3.44GGKK349 pKa = 8.24HH350 pKa = 4.2TVCYY354 pKa = 9.37GQNIPSGYY362 pKa = 9.46IQTTKK367 pKa = 10.87GSDD370 pKa = 3.87CNDD373 pKa = 2.85NDD375 pKa = 4.55ANINPTTVWYY385 pKa = 10.62KK386 pKa = 10.87DD387 pKa = 3.35ADD389 pKa = 3.62NDD391 pKa = 4.49GYY393 pKa = 11.71SDD395 pKa = 3.8GTTKK399 pKa = 8.92TQCDD403 pKa = 3.31QPAGYY408 pKa = 9.59KK409 pKa = 10.36LKK411 pKa = 10.9AQLTATNGDD420 pKa = 4.3CKK422 pKa = 11.18DD423 pKa = 4.15DD424 pKa = 4.65DD425 pKa = 4.08ATIHH429 pKa = 6.62PGAPEE434 pKa = 3.61ICGNGIDD441 pKa = 4.59EE442 pKa = 5.21DD443 pKa = 4.77CDD445 pKa = 4.1SKK447 pKa = 11.74DD448 pKa = 3.62AVCVPTDD455 pKa = 3.29SDD457 pKa = 4.03GDD459 pKa = 4.09GVSDD463 pKa = 6.01NEE465 pKa = 4.44DD466 pKa = 3.67CSPNDD471 pKa = 3.21NKK473 pKa = 10.15VWRR476 pKa = 11.84TVTLYY481 pKa = 11.25ADD483 pKa = 4.38FDD485 pKa = 5.08SDD487 pKa = 3.78GKK489 pKa = 9.87PVAFGSEE496 pKa = 4.09VCIGADD502 pKa = 2.94IPQGYY507 pKa = 9.11SEE509 pKa = 5.4SPGSDD514 pKa = 3.51CDD516 pKa = 5.54DD517 pKa = 3.82NDD519 pKa = 3.41NTVWRR524 pKa = 11.84TAILYY529 pKa = 9.88IDD531 pKa = 3.97SDD533 pKa = 4.05RR534 pKa = 11.84DD535 pKa = 3.85GEE537 pKa = 4.68SVGAGIEE544 pKa = 4.0KK545 pKa = 10.57CIGNDD550 pKa = 2.72IPFGYY555 pKa = 8.87TEE557 pKa = 5.11SPGSDD562 pKa = 3.48CNDD565 pKa = 3.49NNPDD569 pKa = 3.4IYY571 pKa = 10.92HH572 pKa = 7.0GATEE576 pKa = 4.12ICDD579 pKa = 3.84GVDD582 pKa = 3.45NNCDD586 pKa = 3.4GQIDD590 pKa = 4.0EE591 pKa = 5.99GLLFWIYY598 pKa = 10.46PDD600 pKa = 4.36GDD602 pKa = 3.45GDD604 pKa = 4.53GYY606 pKa = 10.2GTEE609 pKa = 3.84EE610 pKa = 4.24GKK612 pKa = 10.29IYY614 pKa = 10.48SCNAPYY620 pKa = 10.55GYY622 pKa = 10.84ADD624 pKa = 4.5RR625 pKa = 11.84NGDD628 pKa = 4.75CKK630 pKa = 11.31DD631 pKa = 4.04DD632 pKa = 5.09DD633 pKa = 3.84NTINPGVEE641 pKa = 4.44EE642 pKa = 4.3ICDD645 pKa = 3.73DD646 pKa = 5.88GIDD649 pKa = 3.79NDD651 pKa = 4.45CDD653 pKa = 4.12GEE655 pKa = 4.05IDD657 pKa = 3.74EE658 pKa = 5.22GCSVSEE664 pKa = 3.8PTEE667 pKa = 4.71FYY669 pKa = 11.1SKK671 pKa = 9.55PTGDD675 pKa = 3.76LHH677 pKa = 7.35NVATWGVNPDD687 pKa = 3.35GSGTQPADD695 pKa = 3.86FGAGKK700 pKa = 8.29TFNLANRR707 pKa = 11.84AGNYY711 pKa = 8.47TMTGNWTVLGTLVNSSGSQLKK732 pKa = 10.43INGYY736 pKa = 6.21TLSLTTLTGAGTLTGSTTSSLIITGTGGGNFGNINFTSGGGMLKK780 pKa = 10.6AFTLNRR786 pKa = 11.84SGTGAAATIGTALAVYY802 pKa = 9.99DD803 pKa = 4.18VLTITSGALTTGGKK817 pKa = 8.08LTLKK821 pKa = 9.52STATNTARR829 pKa = 11.84VAPVTGTISGNVTVEE844 pKa = 3.79RR845 pKa = 11.84YY846 pKa = 9.59IPARR850 pKa = 11.84RR851 pKa = 11.84AWRR854 pKa = 11.84LMNAPVGGTQTINQAWQEE872 pKa = 4.27GVTTASPNPNPAPGYY887 pKa = 7.66GTYY890 pKa = 8.98VTVGSVANGFDD901 pKa = 4.01QNILGQSTSSLKK913 pKa = 10.96SFF915 pKa = 4.16
Molecular weight: 96.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T7QCI3|A0A2T7QCI3_9BACT Peptidase_M56 domain-containing protein OS=Terrimonas sp. NS-102 OX=2171976 GN=DC498_06575 PE=4 SV=1
MM1 pKa = 7.49FLFLIDD7 pKa = 4.05CNKK10 pKa = 9.9GHH12 pKa = 7.45ACRR15 pKa = 11.84SFRR18 pKa = 11.84QPAIFLYY25 pKa = 10.34RR26 pKa = 11.84YY27 pKa = 7.86LQKK30 pKa = 10.33PARR33 pKa = 11.84ARR35 pKa = 11.84DD36 pKa = 3.45LQYY39 pKa = 10.85IQLKK43 pKa = 10.22EE44 pKa = 3.88MLNNGLLSLVFSKK57 pKa = 10.83YY58 pKa = 10.34KK59 pKa = 10.13QVALQKK65 pKa = 10.04RR66 pKa = 11.84GYY68 pKa = 8.03TRR70 pKa = 11.84GVYY73 pKa = 10.47LIISNITFRR82 pKa = 11.84LLHH85 pKa = 5.5NHH87 pKa = 5.4SCRR90 pKa = 11.84SFKK93 pKa = 10.97
MM1 pKa = 7.49FLFLIDD7 pKa = 4.05CNKK10 pKa = 9.9GHH12 pKa = 7.45ACRR15 pKa = 11.84SFRR18 pKa = 11.84QPAIFLYY25 pKa = 10.34RR26 pKa = 11.84YY27 pKa = 7.86LQKK30 pKa = 10.33PARR33 pKa = 11.84ARR35 pKa = 11.84DD36 pKa = 3.45LQYY39 pKa = 10.85IQLKK43 pKa = 10.22EE44 pKa = 3.88MLNNGLLSLVFSKK57 pKa = 10.83YY58 pKa = 10.34KK59 pKa = 10.13QVALQKK65 pKa = 10.04RR66 pKa = 11.84GYY68 pKa = 8.03TRR70 pKa = 11.84GVYY73 pKa = 10.47LIISNITFRR82 pKa = 11.84LLHH85 pKa = 5.5NHH87 pKa = 5.4SCRR90 pKa = 11.84SFKK93 pKa = 10.97
Molecular weight: 11.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1778361 |
16 |
2923 |
354.3 |
39.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.511 ± 0.035 | 0.861 ± 0.011 |
5.302 ± 0.024 | 5.592 ± 0.031 |
4.885 ± 0.023 | 6.835 ± 0.033 |
1.855 ± 0.019 | 7.55 ± 0.031 |
7.222 ± 0.03 | 9.018 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.306 ± 0.017 | 5.715 ± 0.03 |
3.871 ± 0.022 | 3.678 ± 0.019 |
3.93 ± 0.021 | 6.304 ± 0.026 |
5.768 ± 0.033 | 6.195 ± 0.024 |
1.339 ± 0.013 | 4.262 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
