
Gordonia phage Madeline
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Nymbaxtervirinae; Nymphadoravirus; unclassified Nymphadoravirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
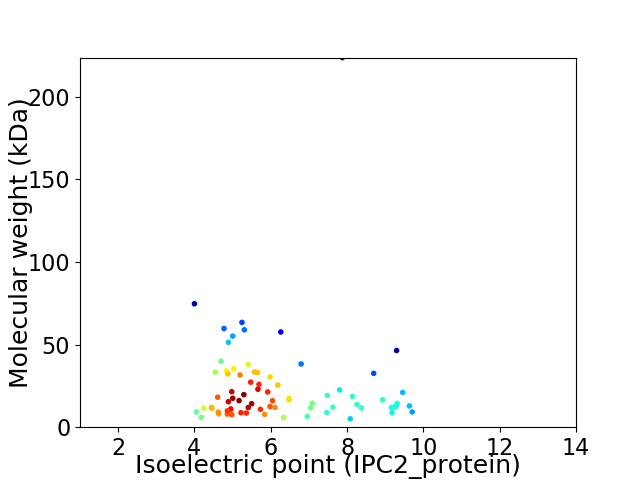
Virtual 2D-PAGE plot for 75 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A514A328|A0A514A328_9CAUD Helix-turn-helix DNA binding domain protein OS=Gordonia phage Madeline OX=2591189 GN=72 PE=4 SV=1
MM1 pKa = 7.17STPLVPDD8 pKa = 3.18WTDD11 pKa = 2.91SDD13 pKa = 4.06IEE15 pKa = 4.34IEE17 pKa = 4.33VPDD20 pKa = 4.07NQVQVIDD27 pKa = 4.08APAVEE32 pKa = 4.43DD33 pKa = 3.94VEE35 pKa = 4.62VLVIQGRR42 pKa = 11.84DD43 pKa = 3.71GQGLQPDD50 pKa = 4.17AAVDD54 pKa = 4.21TYY56 pKa = 12.04ADD58 pKa = 4.33LPDD61 pKa = 4.88DD62 pKa = 5.43LGPNDD67 pKa = 4.05SGQVVYY73 pKa = 10.71VWSDD77 pKa = 2.91KK78 pKa = 11.07LIYY81 pKa = 9.62IWSGTDD87 pKa = 3.02WPAEE91 pKa = 3.95GAGKK95 pKa = 9.87DD96 pKa = 3.4IRR98 pKa = 11.84GMQGDD103 pKa = 4.39PGRR106 pKa = 11.84GIDD109 pKa = 3.62DD110 pKa = 3.73VFVVGTSLVFSMSDD124 pKa = 3.14NTSDD128 pKa = 4.28DD129 pKa = 3.82VPVPAIQQAIDD140 pKa = 3.28SAAAASGSATAADD153 pKa = 4.08TARR156 pKa = 11.84LAAEE160 pKa = 4.66AAASTAGTAASTATTEE176 pKa = 3.77RR177 pKa = 11.84TAAQTARR184 pKa = 11.84TAAEE188 pKa = 4.14AARR191 pKa = 11.84DD192 pKa = 3.65TATNRR197 pKa = 11.84ATAASNSADD206 pKa = 3.11AAATSEE212 pKa = 4.26ANAEE216 pKa = 4.11TSEE219 pKa = 4.54DD220 pKa = 3.61NAATSAAAAATSAGQAANRR239 pKa = 11.84ATDD242 pKa = 3.31ADD244 pKa = 4.03TARR247 pKa = 11.84AAAVLARR254 pKa = 11.84DD255 pKa = 3.72AAAGSATDD263 pKa = 3.64AATSAGSAQTSADD276 pKa = 3.71DD277 pKa = 4.46AGTSAAAAAASAEE290 pKa = 4.09EE291 pKa = 4.16AADD294 pKa = 3.74VVAAGVPDD302 pKa = 3.56ATVTTKK308 pKa = 10.93GGVRR312 pKa = 11.84LAGDD316 pKa = 4.57LGGTWDD322 pKa = 4.35EE323 pKa = 4.25PTVPALAMKK332 pKa = 10.57ADD334 pKa = 4.18LDD336 pKa = 4.64SNGKK340 pKa = 9.07LLSSQMPAQATHH352 pKa = 6.33EE353 pKa = 4.64SVVVTSTAEE362 pKa = 3.9RR363 pKa = 11.84LALTPAQVQPGDD375 pKa = 3.47TAIQLGNPGRR385 pKa = 11.84GTYY388 pKa = 10.03SLQGADD394 pKa = 3.84PSDD397 pKa = 3.83PGSWVLQVAPTDD409 pKa = 3.58AVSSVNGYY417 pKa = 9.61QGIVVLGKK425 pKa = 10.87GDD427 pKa = 3.53VGLGNVDD434 pKa = 3.46NTSDD438 pKa = 3.61LAKK441 pKa = 10.07PISTAAQTALDD452 pKa = 3.98GKK454 pKa = 10.39VDD456 pKa = 3.87EE457 pKa = 4.75VSTANVAYY465 pKa = 7.43GTKK468 pKa = 9.61TGGVQGTWPVTSAATATTLALRR490 pKa = 11.84GTGGTVAVGTATGPTHH506 pKa = 7.37APTKK510 pKa = 9.82QQLDD514 pKa = 3.72DD515 pKa = 4.24GLSGKK520 pKa = 10.37SDD522 pKa = 3.2TGHH525 pKa = 6.61AHH527 pKa = 6.98DD528 pKa = 5.03AAAIATGTLDD538 pKa = 3.78AARR541 pKa = 11.84LPVGTGSTQVAAGNDD556 pKa = 3.49SRR558 pKa = 11.84IVNAVPNSRR567 pKa = 11.84TVTAGTGLSGGGTLDD582 pKa = 3.23VNRR585 pKa = 11.84TLSVLYY591 pKa = 10.92GNTANTATQGNDD603 pKa = 2.58ARR605 pKa = 11.84LSDD608 pKa = 3.77TRR610 pKa = 11.84TPTDD614 pKa = 3.41NTVSTAKK621 pKa = 10.2IQDD624 pKa = 3.75GAVTLAKK631 pKa = 10.32LATAVSVSIQQMIDD645 pKa = 3.08ASVLAAQLVTINAQTGAYY663 pKa = 8.53TLVATDD669 pKa = 3.4ANKK672 pKa = 10.46AVEE675 pKa = 4.54VTSATAVNVTIPTDD689 pKa = 3.34AVNFPIGTVIEE700 pKa = 4.21IDD702 pKa = 3.17QMGAGKK708 pKa = 10.56VSIVGASGVTVKK720 pKa = 10.36PDD722 pKa = 3.11SPTPTTRR729 pKa = 11.84TQYY732 pKa = 10.75SALVLRR738 pKa = 11.84KK739 pKa = 9.31RR740 pKa = 11.84AANWWLVTGDD750 pKa = 3.84LAA752 pKa = 6.13
MM1 pKa = 7.17STPLVPDD8 pKa = 3.18WTDD11 pKa = 2.91SDD13 pKa = 4.06IEE15 pKa = 4.34IEE17 pKa = 4.33VPDD20 pKa = 4.07NQVQVIDD27 pKa = 4.08APAVEE32 pKa = 4.43DD33 pKa = 3.94VEE35 pKa = 4.62VLVIQGRR42 pKa = 11.84DD43 pKa = 3.71GQGLQPDD50 pKa = 4.17AAVDD54 pKa = 4.21TYY56 pKa = 12.04ADD58 pKa = 4.33LPDD61 pKa = 4.88DD62 pKa = 5.43LGPNDD67 pKa = 4.05SGQVVYY73 pKa = 10.71VWSDD77 pKa = 2.91KK78 pKa = 11.07LIYY81 pKa = 9.62IWSGTDD87 pKa = 3.02WPAEE91 pKa = 3.95GAGKK95 pKa = 9.87DD96 pKa = 3.4IRR98 pKa = 11.84GMQGDD103 pKa = 4.39PGRR106 pKa = 11.84GIDD109 pKa = 3.62DD110 pKa = 3.73VFVVGTSLVFSMSDD124 pKa = 3.14NTSDD128 pKa = 4.28DD129 pKa = 3.82VPVPAIQQAIDD140 pKa = 3.28SAAAASGSATAADD153 pKa = 4.08TARR156 pKa = 11.84LAAEE160 pKa = 4.66AAASTAGTAASTATTEE176 pKa = 3.77RR177 pKa = 11.84TAAQTARR184 pKa = 11.84TAAEE188 pKa = 4.14AARR191 pKa = 11.84DD192 pKa = 3.65TATNRR197 pKa = 11.84ATAASNSADD206 pKa = 3.11AAATSEE212 pKa = 4.26ANAEE216 pKa = 4.11TSEE219 pKa = 4.54DD220 pKa = 3.61NAATSAAAAATSAGQAANRR239 pKa = 11.84ATDD242 pKa = 3.31ADD244 pKa = 4.03TARR247 pKa = 11.84AAAVLARR254 pKa = 11.84DD255 pKa = 3.72AAAGSATDD263 pKa = 3.64AATSAGSAQTSADD276 pKa = 3.71DD277 pKa = 4.46AGTSAAAAAASAEE290 pKa = 4.09EE291 pKa = 4.16AADD294 pKa = 3.74VVAAGVPDD302 pKa = 3.56ATVTTKK308 pKa = 10.93GGVRR312 pKa = 11.84LAGDD316 pKa = 4.57LGGTWDD322 pKa = 4.35EE323 pKa = 4.25PTVPALAMKK332 pKa = 10.57ADD334 pKa = 4.18LDD336 pKa = 4.64SNGKK340 pKa = 9.07LLSSQMPAQATHH352 pKa = 6.33EE353 pKa = 4.64SVVVTSTAEE362 pKa = 3.9RR363 pKa = 11.84LALTPAQVQPGDD375 pKa = 3.47TAIQLGNPGRR385 pKa = 11.84GTYY388 pKa = 10.03SLQGADD394 pKa = 3.84PSDD397 pKa = 3.83PGSWVLQVAPTDD409 pKa = 3.58AVSSVNGYY417 pKa = 9.61QGIVVLGKK425 pKa = 10.87GDD427 pKa = 3.53VGLGNVDD434 pKa = 3.46NTSDD438 pKa = 3.61LAKK441 pKa = 10.07PISTAAQTALDD452 pKa = 3.98GKK454 pKa = 10.39VDD456 pKa = 3.87EE457 pKa = 4.75VSTANVAYY465 pKa = 7.43GTKK468 pKa = 9.61TGGVQGTWPVTSAATATTLALRR490 pKa = 11.84GTGGTVAVGTATGPTHH506 pKa = 7.37APTKK510 pKa = 9.82QQLDD514 pKa = 3.72DD515 pKa = 4.24GLSGKK520 pKa = 10.37SDD522 pKa = 3.2TGHH525 pKa = 6.61AHH527 pKa = 6.98DD528 pKa = 5.03AAAIATGTLDD538 pKa = 3.78AARR541 pKa = 11.84LPVGTGSTQVAAGNDD556 pKa = 3.49SRR558 pKa = 11.84IVNAVPNSRR567 pKa = 11.84TVTAGTGLSGGGTLDD582 pKa = 3.23VNRR585 pKa = 11.84TLSVLYY591 pKa = 10.92GNTANTATQGNDD603 pKa = 2.58ARR605 pKa = 11.84LSDD608 pKa = 3.77TRR610 pKa = 11.84TPTDD614 pKa = 3.41NTVSTAKK621 pKa = 10.2IQDD624 pKa = 3.75GAVTLAKK631 pKa = 10.32LATAVSVSIQQMIDD645 pKa = 3.08ASVLAAQLVTINAQTGAYY663 pKa = 8.53TLVATDD669 pKa = 3.4ANKK672 pKa = 10.46AVEE675 pKa = 4.54VTSATAVNVTIPTDD689 pKa = 3.34AVNFPIGTVIEE700 pKa = 4.21IDD702 pKa = 3.17QMGAGKK708 pKa = 10.56VSIVGASGVTVKK720 pKa = 10.36PDD722 pKa = 3.11SPTPTTRR729 pKa = 11.84TQYY732 pKa = 10.75SALVLRR738 pKa = 11.84KK739 pKa = 9.31RR740 pKa = 11.84AANWWLVTGDD750 pKa = 3.84LAA752 pKa = 6.13
Molecular weight: 74.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A514A2W6|A0A514A2W6_9CAUD Membrane protein OS=Gordonia phage Madeline OX=2591189 GN=23 PE=4 SV=1
MM1 pKa = 7.91RR2 pKa = 11.84APLTRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84SMADD14 pKa = 2.86PLARR18 pKa = 11.84FFTQPFTVRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 8.31TGDD32 pKa = 3.38GAVGPIYY39 pKa = 10.44ADD41 pKa = 3.29PVTLRR46 pKa = 11.84GRR48 pKa = 11.84VNATNRR54 pKa = 11.84LVIDD58 pKa = 3.7DD59 pKa = 4.91RR60 pKa = 11.84GNQVLSAAKK69 pKa = 9.23ISMSITEE76 pKa = 4.56DD77 pKa = 4.38DD78 pKa = 4.19IPTGSQARR86 pKa = 11.84VGVGPWRR93 pKa = 11.84TVIADD98 pKa = 3.51SRR100 pKa = 11.84HH101 pKa = 4.94VGGFHH106 pKa = 7.62KK107 pKa = 11.05SPDD110 pKa = 3.72YY111 pKa = 11.29YY112 pKa = 11.39SIDD115 pKa = 3.74LNN117 pKa = 3.96
MM1 pKa = 7.91RR2 pKa = 11.84APLTRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84SMADD14 pKa = 2.86PLARR18 pKa = 11.84FFTQPFTVRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 8.31TGDD32 pKa = 3.38GAVGPIYY39 pKa = 10.44ADD41 pKa = 3.29PVTLRR46 pKa = 11.84GRR48 pKa = 11.84VNATNRR54 pKa = 11.84LVIDD58 pKa = 3.7DD59 pKa = 4.91RR60 pKa = 11.84GNQVLSAAKK69 pKa = 9.23ISMSITEE76 pKa = 4.56DD77 pKa = 4.38DD78 pKa = 4.19IPTGSQARR86 pKa = 11.84VGVGPWRR93 pKa = 11.84TVIADD98 pKa = 3.51SRR100 pKa = 11.84HH101 pKa = 4.94VGGFHH106 pKa = 7.62KK107 pKa = 11.05SPDD110 pKa = 3.72YY111 pKa = 11.29YY112 pKa = 11.39SIDD115 pKa = 3.74LNN117 pKa = 3.96
Molecular weight: 12.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16941 |
47 |
2146 |
225.9 |
24.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.859 ± 0.676 | 0.697 ± 0.15 |
7.066 ± 0.375 | 5.637 ± 0.285 |
2.574 ± 0.194 | 8.317 ± 0.344 |
2.007 ± 0.206 | 4.48 ± 0.16 |
3.288 ± 0.197 | 7.745 ± 0.231 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.119 ± 0.178 | 2.904 ± 0.206 |
5.82 ± 0.254 | 3.601 ± 0.255 |
7.739 ± 0.45 | 5.661 ± 0.216 |
7.278 ± 0.343 | 7.131 ± 0.227 |
1.913 ± 0.152 | 2.166 ± 0.152 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
