
Chytriomyces confervae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Chytridiomycota; Chytridiomycota incertae sedis; Chytridiomycetes; Chytridiales; Chytriomycetaceae; Chytriomyces
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
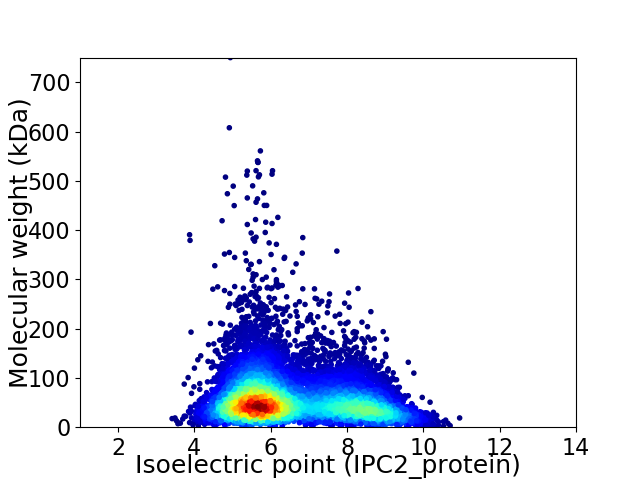
Virtual 2D-PAGE plot for 10661 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A507FKB3|A0A507FKB3_9FUNG NTP_transf_2 domain-containing protein OS=Chytriomyces confervae OX=246404 GN=CcCBS67573_g02879 PE=4 SV=1
MM1 pKa = 7.28FATSANLDD9 pKa = 3.87TRR11 pKa = 11.84PSAPDD16 pKa = 3.34SPSEE20 pKa = 3.86QASVKK25 pKa = 9.84TSFGSYY31 pKa = 9.73IIRR34 pKa = 11.84DD35 pKa = 3.6VRR37 pKa = 11.84FVIATACDD45 pKa = 3.59ISPGPSTKK53 pKa = 10.1NRR55 pKa = 11.84LILPKK60 pKa = 9.21PASVFYY66 pKa = 11.19SPITTCITLYY76 pKa = 9.66GTTHH80 pKa = 6.61PRR82 pKa = 11.84SKK84 pKa = 11.1NLLLEE89 pKa = 4.3MIFNKK94 pKa = 9.92IVATAILFAYY104 pKa = 10.32ASIAFPANIAPPSEE118 pKa = 4.28VAPGGAGTSIPSAEE132 pKa = 4.18PQEE135 pKa = 4.04ITPTSFAPVPSPLLDD150 pKa = 3.35TAFPPVANDD159 pKa = 3.22INGLCGGGSAFKK171 pKa = 10.99NPLACKK177 pKa = 9.86PGLVCAKK184 pKa = 10.12QVGSSDD190 pKa = 3.79DD191 pKa = 3.63TLGTCQIPNVSDD203 pKa = 3.25VGGSCGGEE211 pKa = 3.96VFNAATCAVGLVCISDD227 pKa = 3.84AADD230 pKa = 3.11ATAPGTCQIVVATPTPTSEE249 pKa = 3.92PVGLGGLVTGTEE261 pKa = 4.2EE262 pKa = 4.14VVATGGIVGTEE273 pKa = 3.82PVGTPSEE280 pKa = 4.42TVAGGGIVGTEE291 pKa = 3.91LVEE294 pKa = 3.89PTIAATSEE302 pKa = 4.38TTVSAPVPSSTTTTSKK318 pKa = 7.98TTTTSTASSIVSTVFGEE335 pKa = 4.27NFPPALTSFTTTSEE349 pKa = 3.86EE350 pKa = 3.98LAPPSPSNGAASVLPTLGTEE370 pKa = 4.08PAEE373 pKa = 4.36PPTPSVAPAGTEE385 pKa = 3.91PVTEE389 pKa = 4.43TPSPAGPSTPTEE401 pKa = 4.25APASSTTSEE410 pKa = 4.3EE411 pKa = 4.28APPTPSNPAAGTQPVPEE428 pKa = 4.74PSSTSATTATSEE440 pKa = 4.63EE441 pKa = 4.34IPPTPSIGEE450 pKa = 4.28AEE452 pKa = 4.3TEE454 pKa = 4.22TLDD457 pKa = 3.81EE458 pKa = 4.34LTISPLTGNPPTLPTTTTSEE478 pKa = 4.2GAPPSPSVPAAEE490 pKa = 4.31TEE492 pKa = 4.42PVSSADD498 pKa = 3.97KK499 pKa = 10.8ISTTTDD505 pKa = 2.81TTFIEE510 pKa = 4.73VPPSLSVPAAEE521 pKa = 4.65TEE523 pKa = 4.32PVPPTDD529 pKa = 3.47SSSATSSEE537 pKa = 3.83EE538 pKa = 3.83TTTVIEE544 pKa = 4.45VPPSPSVPAAEE555 pKa = 4.46TEE557 pKa = 4.27PVPPADD563 pKa = 3.65TSSAATTADD572 pKa = 3.67KK573 pKa = 10.65PPQEE577 pKa = 4.4SSSVTDD583 pKa = 3.2SSAPAEE589 pKa = 4.4PNSSEE594 pKa = 4.27STSDD598 pKa = 2.79TTAAVAEE605 pKa = 4.66PEE607 pKa = 4.49TTTQAPTSAAEE618 pKa = 3.77ITTAAEE624 pKa = 4.11ATTTAAEE631 pKa = 4.26IVSSEE636 pKa = 4.28EE637 pKa = 3.89PSTTTSDD644 pKa = 3.21SAIFIPEE651 pKa = 4.2GPTEE655 pKa = 4.01EE656 pKa = 4.75SSTSVEE662 pKa = 4.11TTTASDD668 pKa = 5.43DD669 pKa = 3.91IPPSPSDD676 pKa = 3.39GAGDD680 pKa = 3.4IVSTTTAAAIDD691 pKa = 3.88VSLEE695 pKa = 3.99TSEE698 pKa = 4.88SQVAPSPSTSEE709 pKa = 4.05EE710 pKa = 4.41VPTTTTTTSVEE721 pKa = 4.35SSVTTADD728 pKa = 3.2AGPRR732 pKa = 11.84ALTLPEE738 pKa = 4.68AYY740 pKa = 10.41SLVTRR745 pKa = 11.84EE746 pKa = 4.24LSSLPSCLIGCLNPDD761 pKa = 3.35HH762 pKa = 7.05SSSVTDD768 pKa = 3.49DD769 pKa = 3.56TANDD773 pKa = 3.68LCIDD777 pKa = 4.03AFGNDD782 pKa = 3.37PFALVMCIVSDD793 pKa = 3.92CTGGDD798 pKa = 3.28FDD800 pKa = 5.01VVINALTDD808 pKa = 3.48PTITEE813 pKa = 4.37GLSNACNVIYY823 pKa = 10.58ASIPEE828 pKa = 4.41TISTSFEE835 pKa = 3.92SSSAAEE841 pKa = 4.06PTTTTTAASVAAAPVSPIFSSSTWSMFAAAPVKK874 pKa = 10.69NGVNGTCGGYY884 pKa = 10.39AADD887 pKa = 5.52AATCLPGLKK896 pKa = 9.88CVSKK900 pKa = 10.81SVNEE904 pKa = 4.05VGTCQKK910 pKa = 10.56PVVNDD915 pKa = 3.24VGQTCGGSITNNPASCAGSLACMPNFIPGLPGTCQMPVEE954 pKa = 4.5PSPLTSLSSSSIVVRR969 pKa = 11.84PTTTKK974 pKa = 9.45KK975 pKa = 8.64HH976 pKa = 5.32KK977 pKa = 9.68KK978 pKa = 7.92YY979 pKa = 10.69RR980 pKa = 11.84MVIKK984 pKa = 9.47TVEE987 pKa = 4.3CTTTTVTTTTSTQYY1001 pKa = 11.12AIQSSSPP1008 pKa = 3.22
MM1 pKa = 7.28FATSANLDD9 pKa = 3.87TRR11 pKa = 11.84PSAPDD16 pKa = 3.34SPSEE20 pKa = 3.86QASVKK25 pKa = 9.84TSFGSYY31 pKa = 9.73IIRR34 pKa = 11.84DD35 pKa = 3.6VRR37 pKa = 11.84FVIATACDD45 pKa = 3.59ISPGPSTKK53 pKa = 10.1NRR55 pKa = 11.84LILPKK60 pKa = 9.21PASVFYY66 pKa = 11.19SPITTCITLYY76 pKa = 9.66GTTHH80 pKa = 6.61PRR82 pKa = 11.84SKK84 pKa = 11.1NLLLEE89 pKa = 4.3MIFNKK94 pKa = 9.92IVATAILFAYY104 pKa = 10.32ASIAFPANIAPPSEE118 pKa = 4.28VAPGGAGTSIPSAEE132 pKa = 4.18PQEE135 pKa = 4.04ITPTSFAPVPSPLLDD150 pKa = 3.35TAFPPVANDD159 pKa = 3.22INGLCGGGSAFKK171 pKa = 10.99NPLACKK177 pKa = 9.86PGLVCAKK184 pKa = 10.12QVGSSDD190 pKa = 3.79DD191 pKa = 3.63TLGTCQIPNVSDD203 pKa = 3.25VGGSCGGEE211 pKa = 3.96VFNAATCAVGLVCISDD227 pKa = 3.84AADD230 pKa = 3.11ATAPGTCQIVVATPTPTSEE249 pKa = 3.92PVGLGGLVTGTEE261 pKa = 4.2EE262 pKa = 4.14VVATGGIVGTEE273 pKa = 3.82PVGTPSEE280 pKa = 4.42TVAGGGIVGTEE291 pKa = 3.91LVEE294 pKa = 3.89PTIAATSEE302 pKa = 4.38TTVSAPVPSSTTTTSKK318 pKa = 7.98TTTTSTASSIVSTVFGEE335 pKa = 4.27NFPPALTSFTTTSEE349 pKa = 3.86EE350 pKa = 3.98LAPPSPSNGAASVLPTLGTEE370 pKa = 4.08PAEE373 pKa = 4.36PPTPSVAPAGTEE385 pKa = 3.91PVTEE389 pKa = 4.43TPSPAGPSTPTEE401 pKa = 4.25APASSTTSEE410 pKa = 4.3EE411 pKa = 4.28APPTPSNPAAGTQPVPEE428 pKa = 4.74PSSTSATTATSEE440 pKa = 4.63EE441 pKa = 4.34IPPTPSIGEE450 pKa = 4.28AEE452 pKa = 4.3TEE454 pKa = 4.22TLDD457 pKa = 3.81EE458 pKa = 4.34LTISPLTGNPPTLPTTTTSEE478 pKa = 4.2GAPPSPSVPAAEE490 pKa = 4.31TEE492 pKa = 4.42PVSSADD498 pKa = 3.97KK499 pKa = 10.8ISTTTDD505 pKa = 2.81TTFIEE510 pKa = 4.73VPPSLSVPAAEE521 pKa = 4.65TEE523 pKa = 4.32PVPPTDD529 pKa = 3.47SSSATSSEE537 pKa = 3.83EE538 pKa = 3.83TTTVIEE544 pKa = 4.45VPPSPSVPAAEE555 pKa = 4.46TEE557 pKa = 4.27PVPPADD563 pKa = 3.65TSSAATTADD572 pKa = 3.67KK573 pKa = 10.65PPQEE577 pKa = 4.4SSSVTDD583 pKa = 3.2SSAPAEE589 pKa = 4.4PNSSEE594 pKa = 4.27STSDD598 pKa = 2.79TTAAVAEE605 pKa = 4.66PEE607 pKa = 4.49TTTQAPTSAAEE618 pKa = 3.77ITTAAEE624 pKa = 4.11ATTTAAEE631 pKa = 4.26IVSSEE636 pKa = 4.28EE637 pKa = 3.89PSTTTSDD644 pKa = 3.21SAIFIPEE651 pKa = 4.2GPTEE655 pKa = 4.01EE656 pKa = 4.75SSTSVEE662 pKa = 4.11TTTASDD668 pKa = 5.43DD669 pKa = 3.91IPPSPSDD676 pKa = 3.39GAGDD680 pKa = 3.4IVSTTTAAAIDD691 pKa = 3.88VSLEE695 pKa = 3.99TSEE698 pKa = 4.88SQVAPSPSTSEE709 pKa = 4.05EE710 pKa = 4.41VPTTTTTTSVEE721 pKa = 4.35SSVTTADD728 pKa = 3.2AGPRR732 pKa = 11.84ALTLPEE738 pKa = 4.68AYY740 pKa = 10.41SLVTRR745 pKa = 11.84EE746 pKa = 4.24LSSLPSCLIGCLNPDD761 pKa = 3.35HH762 pKa = 7.05SSSVTDD768 pKa = 3.49DD769 pKa = 3.56TANDD773 pKa = 3.68LCIDD777 pKa = 4.03AFGNDD782 pKa = 3.37PFALVMCIVSDD793 pKa = 3.92CTGGDD798 pKa = 3.28FDD800 pKa = 5.01VVINALTDD808 pKa = 3.48PTITEE813 pKa = 4.37GLSNACNVIYY823 pKa = 10.58ASIPEE828 pKa = 4.41TISTSFEE835 pKa = 3.92SSSAAEE841 pKa = 4.06PTTTTTAASVAAAPVSPIFSSSTWSMFAAAPVKK874 pKa = 10.69NGVNGTCGGYY884 pKa = 10.39AADD887 pKa = 5.52AATCLPGLKK896 pKa = 9.88CVSKK900 pKa = 10.81SVNEE904 pKa = 4.05VGTCQKK910 pKa = 10.56PVVNDD915 pKa = 3.24VGQTCGGSITNNPASCAGSLACMPNFIPGLPGTCQMPVEE954 pKa = 4.5PSPLTSLSSSSIVVRR969 pKa = 11.84PTTTKK974 pKa = 9.45KK975 pKa = 8.64HH976 pKa = 5.32KK977 pKa = 9.68KK978 pKa = 7.92YY979 pKa = 10.69RR980 pKa = 11.84MVIKK984 pKa = 9.47TVEE987 pKa = 4.3CTTTTVTTTTSTQYY1001 pKa = 11.12AIQSSSPP1008 pKa = 3.22
Molecular weight: 100.75 kDa
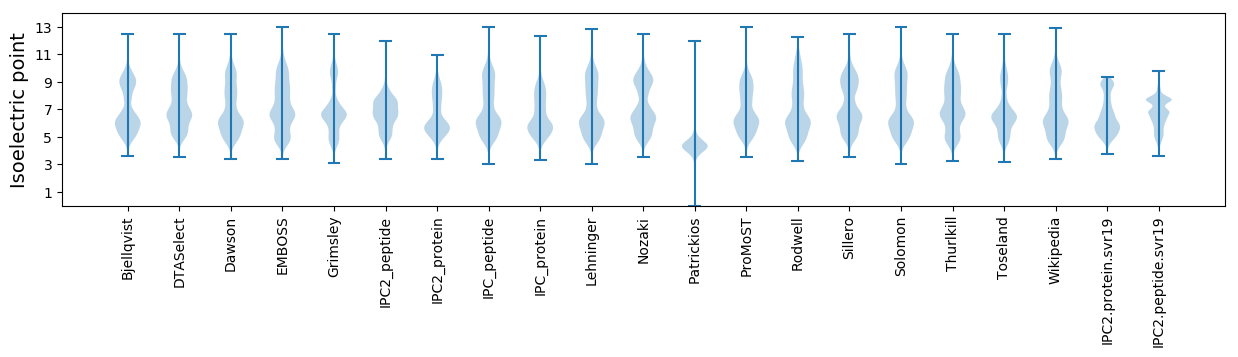
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A507D400|A0A507D400_9FUNG Pirin domain-containing protein OS=Chytriomyces confervae OX=246404 GN=CcCBS67573_g10330 PE=3 SV=1
MM1 pKa = 7.54SSARR5 pKa = 11.84LTGPIPGSFGNMINLVEE22 pKa = 5.81LRR24 pKa = 11.84LQMNKK29 pKa = 10.33LLGTVLVIACNSRR42 pKa = 11.84PSNAKK47 pKa = 9.24RR48 pKa = 11.84INFRR52 pKa = 11.84RR53 pKa = 11.84PTTCNASAEE62 pKa = 4.23RR63 pKa = 11.84IEE65 pKa = 4.5MGDD68 pKa = 3.49NQLTPIVDD76 pKa = 3.24IDD78 pKa = 3.57SDD80 pKa = 3.76RR81 pKa = 11.84TSGYY85 pKa = 9.48IYY87 pKa = 9.24PFPSVSLDD95 pKa = 3.56LQSPNAGSNQLQEE108 pKa = 5.34DD109 pKa = 4.4SFADD113 pKa = 4.37TIDD116 pKa = 3.56SDD118 pKa = 4.75SMFTSRR124 pKa = 11.84LGLSRR129 pKa = 11.84TQLSRR134 pKa = 11.84KK135 pKa = 9.46LPNCSKK141 pKa = 10.76KK142 pKa = 10.65SATHH146 pKa = 5.77SPQRR150 pKa = 11.84QQSRR154 pKa = 11.84WSVSHH159 pKa = 6.63RR160 pKa = 11.84NWRR163 pKa = 11.84LRR165 pKa = 11.84KK166 pKa = 8.82IGKK169 pKa = 9.14RR170 pKa = 11.84RR171 pKa = 3.3
MM1 pKa = 7.54SSARR5 pKa = 11.84LTGPIPGSFGNMINLVEE22 pKa = 5.81LRR24 pKa = 11.84LQMNKK29 pKa = 10.33LLGTVLVIACNSRR42 pKa = 11.84PSNAKK47 pKa = 9.24RR48 pKa = 11.84INFRR52 pKa = 11.84RR53 pKa = 11.84PTTCNASAEE62 pKa = 4.23RR63 pKa = 11.84IEE65 pKa = 4.5MGDD68 pKa = 3.49NQLTPIVDD76 pKa = 3.24IDD78 pKa = 3.57SDD80 pKa = 3.76RR81 pKa = 11.84TSGYY85 pKa = 9.48IYY87 pKa = 9.24PFPSVSLDD95 pKa = 3.56LQSPNAGSNQLQEE108 pKa = 5.34DD109 pKa = 4.4SFADD113 pKa = 4.37TIDD116 pKa = 3.56SDD118 pKa = 4.75SMFTSRR124 pKa = 11.84LGLSRR129 pKa = 11.84TQLSRR134 pKa = 11.84KK135 pKa = 9.46LPNCSKK141 pKa = 10.76KK142 pKa = 10.65SATHH146 pKa = 5.77SPQRR150 pKa = 11.84QQSRR154 pKa = 11.84WSVSHH159 pKa = 6.63RR160 pKa = 11.84NWRR163 pKa = 11.84LRR165 pKa = 11.84KK166 pKa = 8.82IGKK169 pKa = 9.14RR170 pKa = 11.84RR171 pKa = 3.3
Molecular weight: 19.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5997939 |
10 |
6810 |
562.6 |
61.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.948 ± 0.02 | 1.337 ± 0.009 |
5.37 ± 0.016 | 5.826 ± 0.026 |
4.059 ± 0.014 | 6.246 ± 0.022 |
2.304 ± 0.011 | 5.087 ± 0.016 |
5.394 ± 0.022 | 9.224 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.008 | 4.235 ± 0.012 |
5.138 ± 0.023 | 3.998 ± 0.018 |
5.055 ± 0.017 | 8.957 ± 0.032 |
5.937 ± 0.025 | 6.8 ± 0.024 |
1.111 ± 0.007 | 2.558 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
