
Aliiglaciecola lipolytica E3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Aliiglaciecola; Aliiglaciecola lipolytica
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
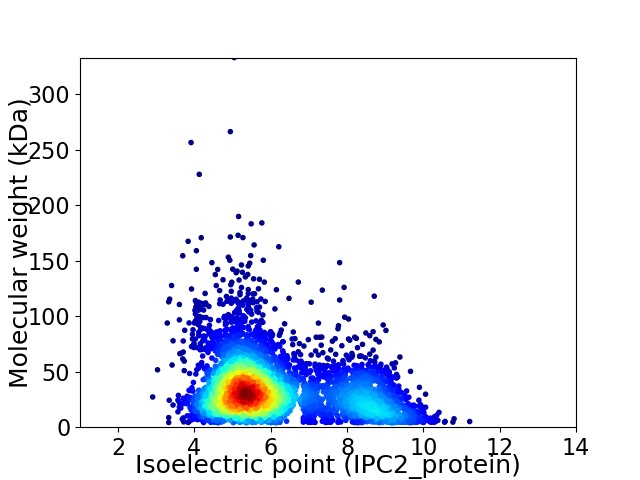
Virtual 2D-PAGE plot for 4327 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K6XUX9|K6XUX9_9ALTE Ubiquinone biosynthesis accessory factor UbiK OS=Aliiglaciecola lipolytica E3 OX=1127673 GN=ubiK PE=3 SV=1
MM1 pKa = 7.46KK2 pKa = 10.05IKK4 pKa = 10.83YY5 pKa = 7.68PALLALVCAPLAHH18 pKa = 6.97ADD20 pKa = 3.41VFINEE25 pKa = 3.75FHH27 pKa = 7.01YY28 pKa = 11.22DD29 pKa = 3.4NSSTDD34 pKa = 3.0SGEE37 pKa = 4.34AIEE40 pKa = 4.4IAGTAGTDD48 pKa = 3.38LSGWSLVLYY57 pKa = 10.06NGSNGSVYY65 pKa = 10.11DD66 pKa = 3.79TTALSGSIPDD76 pKa = 3.65QQNGMGTVFFSYY88 pKa = 10.25PSNGIQNGPDD98 pKa = 3.57AIALVDD104 pKa = 4.32DD105 pKa = 4.69SNSVIQFISYY115 pKa = 10.18EE116 pKa = 4.34GTFSASGGPANGMTSVNIGVSEE138 pKa = 4.2SGSTNVGDD146 pKa = 4.15SLQLSGTGSAYY157 pKa = 10.4ADD159 pKa = 4.04FAWQSAAANTFGAINNDD176 pKa = 3.09QSFSANDD183 pKa = 3.7GGGEE187 pKa = 4.26DD188 pKa = 4.76GGGDD192 pKa = 3.64DD193 pKa = 4.31SGSLEE198 pKa = 4.07GVCFNCPDD206 pKa = 3.91LDD208 pKa = 3.91KK209 pKa = 11.51VADD212 pKa = 3.79ASEE215 pKa = 4.5FDD217 pKa = 3.76DD218 pKa = 3.99ASYY221 pKa = 10.96YY222 pKa = 10.82AAAKK226 pKa = 9.62TEE228 pKa = 3.74IDD230 pKa = 3.81MGSSSAIIKK239 pKa = 10.12SALNGIITNDD249 pKa = 3.4HH250 pKa = 6.35RR251 pKa = 11.84VLTYY255 pKa = 10.7DD256 pKa = 3.78QVWTALTEE264 pKa = 4.22SDD266 pKa = 4.24EE267 pKa = 5.34DD268 pKa = 3.93PANSDD273 pKa = 3.5NVLLIYY279 pKa = 10.47KK280 pKa = 9.69GISLPKK286 pKa = 9.66LSNGSGEE293 pKa = 4.26QSTDD297 pKa = 3.08PDD299 pKa = 2.98NWNRR303 pKa = 11.84EE304 pKa = 4.4HH305 pKa = 6.31VWPNSHH311 pKa = 6.47GFPDD315 pKa = 4.18RR316 pKa = 11.84PFEE319 pKa = 4.47AYY321 pKa = 9.4TDD323 pKa = 3.46IHH325 pKa = 6.55HH326 pKa = 7.07LRR328 pKa = 11.84PSDD331 pKa = 3.09ISVNGSRR338 pKa = 11.84GNLDD342 pKa = 3.91FDD344 pKa = 4.82NSDD347 pKa = 3.61APLDD351 pKa = 3.83EE352 pKa = 5.23APSNRR357 pKa = 11.84IDD359 pKa = 3.37SDD361 pKa = 3.68SFEE364 pKa = 4.72PRR366 pKa = 11.84DD367 pKa = 3.61EE368 pKa = 4.35VKK370 pKa = 11.11GDD372 pKa = 3.37VARR375 pKa = 11.84MMFYY379 pKa = 9.97MDD381 pKa = 3.22TRR383 pKa = 11.84YY384 pKa = 10.51EE385 pKa = 4.22GLGDD389 pKa = 3.74ATPDD393 pKa = 3.36LVLSNGLTSAGQPQLGRR410 pKa = 11.84LCRR413 pKa = 11.84LIEE416 pKa = 3.92WHH418 pKa = 6.58NADD421 pKa = 4.07PVDD424 pKa = 3.44AAEE427 pKa = 4.2QQRR430 pKa = 11.84HH431 pKa = 4.3DD432 pKa = 4.29TIYY435 pKa = 10.52EE436 pKa = 3.96YY437 pKa = 10.71QGNRR441 pKa = 11.84NPFVDD446 pKa = 3.48HH447 pKa = 6.97PEE449 pKa = 3.95WVSLLFTAEE458 pKa = 3.84VCGDD462 pKa = 3.84DD463 pKa = 5.92DD464 pKa = 4.74GGEE467 pKa = 4.29DD468 pKa = 4.61GGDD471 pKa = 3.56DD472 pKa = 4.58GEE474 pKa = 5.33DD475 pKa = 3.49SGNVGAVEE483 pKa = 4.1ANALLITEE491 pKa = 4.76IMQNPDD497 pKa = 3.88TISDD501 pKa = 3.89ANGEE505 pKa = 4.08WFEE508 pKa = 4.14VLNTSTQVINLNGWTIRR525 pKa = 11.84DD526 pKa = 3.37NDD528 pKa = 3.35FDD530 pKa = 5.04SFVVDD535 pKa = 3.04QDD537 pKa = 3.57VFIGVGEE544 pKa = 4.08YY545 pKa = 10.81AVLGKK550 pKa = 10.09NADD553 pKa = 3.46TSVNGGITLAYY564 pKa = 10.13AYY566 pKa = 9.42GGSMALANGSDD577 pKa = 3.87EE578 pKa = 4.83LVLVDD583 pKa = 4.04PDD585 pKa = 4.0GNIVDD590 pKa = 5.22EE591 pKa = 4.26ITYY594 pKa = 10.61DD595 pKa = 3.67GGPDD599 pKa = 3.57FPDD602 pKa = 3.37PTGASMYY609 pKa = 10.92LVDD612 pKa = 6.26LYY614 pKa = 11.62GDD616 pKa = 3.75NNVGSKK622 pKa = 8.47WAADD626 pKa = 3.37TTNFYY631 pKa = 11.28DD632 pKa = 4.13SNNSGTPGTGDD643 pKa = 2.85ASFNLVITEE652 pKa = 4.07IMQNPRR658 pKa = 11.84AVGDD662 pKa = 3.73SAGEE666 pKa = 3.85YY667 pKa = 10.71VEE669 pKa = 4.31ILNTGVIAANLNGWVFRR686 pKa = 11.84DD687 pKa = 3.55NDD689 pKa = 4.32SEE691 pKa = 4.56SHH693 pKa = 6.58TIQGDD698 pKa = 4.06VYY700 pKa = 10.98VPAGTYY706 pKa = 10.34AVLTNNSDD714 pKa = 3.38SSSNGGFEE722 pKa = 5.45ALYY725 pKa = 9.94QYY727 pKa = 11.2SGVFIANGADD737 pKa = 3.12EE738 pKa = 5.25VVLQRR743 pKa = 11.84PDD745 pKa = 3.06GTIEE749 pKa = 5.7DD750 pKa = 3.51IVEE753 pKa = 4.06YY754 pKa = 10.69DD755 pKa = 3.76GGPTFPDD762 pKa = 3.51PNGASMTLISASLDD776 pKa = 3.51NNVGEE781 pKa = 4.27NWVEE785 pKa = 4.0EE786 pKa = 4.32TTATYY791 pKa = 11.28GDD793 pKa = 4.36GDD795 pKa = 4.12FGTPGSGPDD804 pKa = 3.14GKK806 pKa = 9.08TFGGGDD812 pKa = 4.16DD813 pKa = 4.41GPGDD817 pKa = 4.08VDD819 pKa = 4.44PEE821 pKa = 4.2IGVCTDD827 pKa = 3.57PATLISTIQGNDD839 pKa = 3.17FASLLDD845 pKa = 3.99GQEE848 pKa = 4.61HH849 pKa = 5.83IVEE852 pKa = 4.25AVVTSTLTDD861 pKa = 3.74FGGFYY866 pKa = 10.52LQEE869 pKa = 3.8EE870 pKa = 4.61DD871 pKa = 4.66ADD873 pKa = 3.93VDD875 pKa = 3.88ADD877 pKa = 3.91PATSEE882 pKa = 4.07GLYY885 pKa = 10.57VSYY888 pKa = 10.86FDD890 pKa = 5.88ANALPAQGTVVRR902 pKa = 11.84VMGTVAEE909 pKa = 4.33NFGRR913 pKa = 11.84TQLNVSSDD921 pKa = 3.66LVEE924 pKa = 4.97CGTDD928 pKa = 3.25TVSATTLVLPFSSALEE944 pKa = 4.18AEE946 pKa = 4.61SLEE949 pKa = 4.0NMLVVNEE956 pKa = 4.48SPLTVTNTYY965 pKa = 8.69TLARR969 pKa = 11.84FGEE972 pKa = 4.45VGLSFGRR979 pKa = 11.84LFNPTNVYY987 pKa = 10.88APLSPEE993 pKa = 4.12AVDD996 pKa = 5.36LAAQNALNYY1005 pKa = 10.08IIMDD1009 pKa = 4.73DD1010 pKa = 3.98GLDD1013 pKa = 3.53VQNPEE1018 pKa = 3.96TVIYY1022 pKa = 7.53PTGNLSAANTLRR1034 pKa = 11.84TGDD1037 pKa = 3.55TVVNIKK1043 pKa = 10.41GALDD1047 pKa = 3.44YY1048 pKa = 11.17SFSNFRR1054 pKa = 11.84IHH1056 pKa = 7.52PIEE1059 pKa = 4.43TPTIIHH1065 pKa = 5.82SNEE1068 pKa = 3.81RR1069 pKa = 11.84EE1070 pKa = 3.91PAPSITRR1077 pKa = 11.84GNLTVASLNVLNLFNGDD1094 pKa = 3.41GQGGGFPTEE1103 pKa = 4.18RR1104 pKa = 11.84GADD1107 pKa = 3.33SLFEE1111 pKa = 4.05YY1112 pKa = 10.21EE1113 pKa = 4.06RR1114 pKa = 11.84QIVKK1118 pKa = 8.95TVEE1121 pKa = 4.62AISTMDD1127 pKa = 3.62ADD1129 pKa = 3.41IVGLMEE1135 pKa = 4.75IEE1137 pKa = 3.66NDD1139 pKa = 3.61GFGEE1143 pKa = 4.24FSMVAEE1149 pKa = 4.28LTNRR1153 pKa = 11.84LNEE1156 pKa = 3.97VMGEE1160 pKa = 4.04GTYY1163 pKa = 11.21DD1164 pKa = 3.45FVSYY1168 pKa = 10.56SSEE1171 pKa = 3.72IGTDD1175 pKa = 4.1AIAVALLYY1183 pKa = 10.68KK1184 pKa = 9.82PASVTLDD1191 pKa = 3.13GDD1193 pKa = 3.71VKK1195 pKa = 11.16INFDD1199 pKa = 4.36SIFNRR1204 pKa = 11.84PPVAQSFTTLNGAGITVVVNHH1225 pKa = 6.64FKK1227 pKa = 11.19SKK1229 pKa = 10.25GCGSASGDD1237 pKa = 4.02DD1238 pKa = 3.74TDD1240 pKa = 4.5QGDD1243 pKa = 4.04GQGCYY1248 pKa = 8.24NAKK1251 pKa = 8.79RR1252 pKa = 11.84TQQSLSLASWLASEE1266 pKa = 5.05EE1267 pKa = 4.16SLSTKK1272 pKa = 10.43EE1273 pKa = 3.74NVLIIGDD1280 pKa = 4.23LNAYY1284 pKa = 9.97AKK1286 pKa = 9.71EE1287 pKa = 4.06DD1288 pKa = 4.53PIAALEE1294 pKa = 4.22GQGFVNLVEE1303 pKa = 4.32TFQGAEE1309 pKa = 3.92AYY1311 pKa = 10.2SYY1313 pKa = 9.83TFSGEE1318 pKa = 3.73FGYY1321 pKa = 10.78LDD1323 pKa = 3.55HH1324 pKa = 7.28ALASSSLAAQAVDD1337 pKa = 3.69TIEE1340 pKa = 3.61WHH1342 pKa = 6.61INADD1346 pKa = 3.53EE1347 pKa = 4.49PFALDD1352 pKa = 3.4YY1353 pKa = 11.21NVEE1356 pKa = 4.45FKK1358 pKa = 11.08SDD1360 pKa = 3.19AQVNDD1365 pKa = 5.39FYY1367 pKa = 11.61ASDD1370 pKa = 3.66VFRR1373 pKa = 11.84VSDD1376 pKa = 4.17HH1377 pKa = 7.21DD1378 pKa = 3.72PVMMSFEE1385 pKa = 4.26LASPAVEE1392 pKa = 4.12GDD1394 pKa = 2.67IDD1396 pKa = 4.46GDD1398 pKa = 3.63MDD1400 pKa = 3.94VDD1402 pKa = 4.17YY1403 pKa = 11.75NDD1405 pKa = 3.49MRR1407 pKa = 11.84ALMNLIRR1414 pKa = 11.84AGQATLEE1421 pKa = 4.06EE1422 pKa = 4.65HH1423 pKa = 7.33DD1424 pKa = 4.49FNNDD1428 pKa = 2.68GLLNSGDD1435 pKa = 3.63VSALRR1440 pKa = 11.84SMCSRR1445 pKa = 11.84RR1446 pKa = 11.84ACSTRR1451 pKa = 3.02
MM1 pKa = 7.46KK2 pKa = 10.05IKK4 pKa = 10.83YY5 pKa = 7.68PALLALVCAPLAHH18 pKa = 6.97ADD20 pKa = 3.41VFINEE25 pKa = 3.75FHH27 pKa = 7.01YY28 pKa = 11.22DD29 pKa = 3.4NSSTDD34 pKa = 3.0SGEE37 pKa = 4.34AIEE40 pKa = 4.4IAGTAGTDD48 pKa = 3.38LSGWSLVLYY57 pKa = 10.06NGSNGSVYY65 pKa = 10.11DD66 pKa = 3.79TTALSGSIPDD76 pKa = 3.65QQNGMGTVFFSYY88 pKa = 10.25PSNGIQNGPDD98 pKa = 3.57AIALVDD104 pKa = 4.32DD105 pKa = 4.69SNSVIQFISYY115 pKa = 10.18EE116 pKa = 4.34GTFSASGGPANGMTSVNIGVSEE138 pKa = 4.2SGSTNVGDD146 pKa = 4.15SLQLSGTGSAYY157 pKa = 10.4ADD159 pKa = 4.04FAWQSAAANTFGAINNDD176 pKa = 3.09QSFSANDD183 pKa = 3.7GGGEE187 pKa = 4.26DD188 pKa = 4.76GGGDD192 pKa = 3.64DD193 pKa = 4.31SGSLEE198 pKa = 4.07GVCFNCPDD206 pKa = 3.91LDD208 pKa = 3.91KK209 pKa = 11.51VADD212 pKa = 3.79ASEE215 pKa = 4.5FDD217 pKa = 3.76DD218 pKa = 3.99ASYY221 pKa = 10.96YY222 pKa = 10.82AAAKK226 pKa = 9.62TEE228 pKa = 3.74IDD230 pKa = 3.81MGSSSAIIKK239 pKa = 10.12SALNGIITNDD249 pKa = 3.4HH250 pKa = 6.35RR251 pKa = 11.84VLTYY255 pKa = 10.7DD256 pKa = 3.78QVWTALTEE264 pKa = 4.22SDD266 pKa = 4.24EE267 pKa = 5.34DD268 pKa = 3.93PANSDD273 pKa = 3.5NVLLIYY279 pKa = 10.47KK280 pKa = 9.69GISLPKK286 pKa = 9.66LSNGSGEE293 pKa = 4.26QSTDD297 pKa = 3.08PDD299 pKa = 2.98NWNRR303 pKa = 11.84EE304 pKa = 4.4HH305 pKa = 6.31VWPNSHH311 pKa = 6.47GFPDD315 pKa = 4.18RR316 pKa = 11.84PFEE319 pKa = 4.47AYY321 pKa = 9.4TDD323 pKa = 3.46IHH325 pKa = 6.55HH326 pKa = 7.07LRR328 pKa = 11.84PSDD331 pKa = 3.09ISVNGSRR338 pKa = 11.84GNLDD342 pKa = 3.91FDD344 pKa = 4.82NSDD347 pKa = 3.61APLDD351 pKa = 3.83EE352 pKa = 5.23APSNRR357 pKa = 11.84IDD359 pKa = 3.37SDD361 pKa = 3.68SFEE364 pKa = 4.72PRR366 pKa = 11.84DD367 pKa = 3.61EE368 pKa = 4.35VKK370 pKa = 11.11GDD372 pKa = 3.37VARR375 pKa = 11.84MMFYY379 pKa = 9.97MDD381 pKa = 3.22TRR383 pKa = 11.84YY384 pKa = 10.51EE385 pKa = 4.22GLGDD389 pKa = 3.74ATPDD393 pKa = 3.36LVLSNGLTSAGQPQLGRR410 pKa = 11.84LCRR413 pKa = 11.84LIEE416 pKa = 3.92WHH418 pKa = 6.58NADD421 pKa = 4.07PVDD424 pKa = 3.44AAEE427 pKa = 4.2QQRR430 pKa = 11.84HH431 pKa = 4.3DD432 pKa = 4.29TIYY435 pKa = 10.52EE436 pKa = 3.96YY437 pKa = 10.71QGNRR441 pKa = 11.84NPFVDD446 pKa = 3.48HH447 pKa = 6.97PEE449 pKa = 3.95WVSLLFTAEE458 pKa = 3.84VCGDD462 pKa = 3.84DD463 pKa = 5.92DD464 pKa = 4.74GGEE467 pKa = 4.29DD468 pKa = 4.61GGDD471 pKa = 3.56DD472 pKa = 4.58GEE474 pKa = 5.33DD475 pKa = 3.49SGNVGAVEE483 pKa = 4.1ANALLITEE491 pKa = 4.76IMQNPDD497 pKa = 3.88TISDD501 pKa = 3.89ANGEE505 pKa = 4.08WFEE508 pKa = 4.14VLNTSTQVINLNGWTIRR525 pKa = 11.84DD526 pKa = 3.37NDD528 pKa = 3.35FDD530 pKa = 5.04SFVVDD535 pKa = 3.04QDD537 pKa = 3.57VFIGVGEE544 pKa = 4.08YY545 pKa = 10.81AVLGKK550 pKa = 10.09NADD553 pKa = 3.46TSVNGGITLAYY564 pKa = 10.13AYY566 pKa = 9.42GGSMALANGSDD577 pKa = 3.87EE578 pKa = 4.83LVLVDD583 pKa = 4.04PDD585 pKa = 4.0GNIVDD590 pKa = 5.22EE591 pKa = 4.26ITYY594 pKa = 10.61DD595 pKa = 3.67GGPDD599 pKa = 3.57FPDD602 pKa = 3.37PTGASMYY609 pKa = 10.92LVDD612 pKa = 6.26LYY614 pKa = 11.62GDD616 pKa = 3.75NNVGSKK622 pKa = 8.47WAADD626 pKa = 3.37TTNFYY631 pKa = 11.28DD632 pKa = 4.13SNNSGTPGTGDD643 pKa = 2.85ASFNLVITEE652 pKa = 4.07IMQNPRR658 pKa = 11.84AVGDD662 pKa = 3.73SAGEE666 pKa = 3.85YY667 pKa = 10.71VEE669 pKa = 4.31ILNTGVIAANLNGWVFRR686 pKa = 11.84DD687 pKa = 3.55NDD689 pKa = 4.32SEE691 pKa = 4.56SHH693 pKa = 6.58TIQGDD698 pKa = 4.06VYY700 pKa = 10.98VPAGTYY706 pKa = 10.34AVLTNNSDD714 pKa = 3.38SSSNGGFEE722 pKa = 5.45ALYY725 pKa = 9.94QYY727 pKa = 11.2SGVFIANGADD737 pKa = 3.12EE738 pKa = 5.25VVLQRR743 pKa = 11.84PDD745 pKa = 3.06GTIEE749 pKa = 5.7DD750 pKa = 3.51IVEE753 pKa = 4.06YY754 pKa = 10.69DD755 pKa = 3.76GGPTFPDD762 pKa = 3.51PNGASMTLISASLDD776 pKa = 3.51NNVGEE781 pKa = 4.27NWVEE785 pKa = 4.0EE786 pKa = 4.32TTATYY791 pKa = 11.28GDD793 pKa = 4.36GDD795 pKa = 4.12FGTPGSGPDD804 pKa = 3.14GKK806 pKa = 9.08TFGGGDD812 pKa = 4.16DD813 pKa = 4.41GPGDD817 pKa = 4.08VDD819 pKa = 4.44PEE821 pKa = 4.2IGVCTDD827 pKa = 3.57PATLISTIQGNDD839 pKa = 3.17FASLLDD845 pKa = 3.99GQEE848 pKa = 4.61HH849 pKa = 5.83IVEE852 pKa = 4.25AVVTSTLTDD861 pKa = 3.74FGGFYY866 pKa = 10.52LQEE869 pKa = 3.8EE870 pKa = 4.61DD871 pKa = 4.66ADD873 pKa = 3.93VDD875 pKa = 3.88ADD877 pKa = 3.91PATSEE882 pKa = 4.07GLYY885 pKa = 10.57VSYY888 pKa = 10.86FDD890 pKa = 5.88ANALPAQGTVVRR902 pKa = 11.84VMGTVAEE909 pKa = 4.33NFGRR913 pKa = 11.84TQLNVSSDD921 pKa = 3.66LVEE924 pKa = 4.97CGTDD928 pKa = 3.25TVSATTLVLPFSSALEE944 pKa = 4.18AEE946 pKa = 4.61SLEE949 pKa = 4.0NMLVVNEE956 pKa = 4.48SPLTVTNTYY965 pKa = 8.69TLARR969 pKa = 11.84FGEE972 pKa = 4.45VGLSFGRR979 pKa = 11.84LFNPTNVYY987 pKa = 10.88APLSPEE993 pKa = 4.12AVDD996 pKa = 5.36LAAQNALNYY1005 pKa = 10.08IIMDD1009 pKa = 4.73DD1010 pKa = 3.98GLDD1013 pKa = 3.53VQNPEE1018 pKa = 3.96TVIYY1022 pKa = 7.53PTGNLSAANTLRR1034 pKa = 11.84TGDD1037 pKa = 3.55TVVNIKK1043 pKa = 10.41GALDD1047 pKa = 3.44YY1048 pKa = 11.17SFSNFRR1054 pKa = 11.84IHH1056 pKa = 7.52PIEE1059 pKa = 4.43TPTIIHH1065 pKa = 5.82SNEE1068 pKa = 3.81RR1069 pKa = 11.84EE1070 pKa = 3.91PAPSITRR1077 pKa = 11.84GNLTVASLNVLNLFNGDD1094 pKa = 3.41GQGGGFPTEE1103 pKa = 4.18RR1104 pKa = 11.84GADD1107 pKa = 3.33SLFEE1111 pKa = 4.05YY1112 pKa = 10.21EE1113 pKa = 4.06RR1114 pKa = 11.84QIVKK1118 pKa = 8.95TVEE1121 pKa = 4.62AISTMDD1127 pKa = 3.62ADD1129 pKa = 3.41IVGLMEE1135 pKa = 4.75IEE1137 pKa = 3.66NDD1139 pKa = 3.61GFGEE1143 pKa = 4.24FSMVAEE1149 pKa = 4.28LTNRR1153 pKa = 11.84LNEE1156 pKa = 3.97VMGEE1160 pKa = 4.04GTYY1163 pKa = 11.21DD1164 pKa = 3.45FVSYY1168 pKa = 10.56SSEE1171 pKa = 3.72IGTDD1175 pKa = 4.1AIAVALLYY1183 pKa = 10.68KK1184 pKa = 9.82PASVTLDD1191 pKa = 3.13GDD1193 pKa = 3.71VKK1195 pKa = 11.16INFDD1199 pKa = 4.36SIFNRR1204 pKa = 11.84PPVAQSFTTLNGAGITVVVNHH1225 pKa = 6.64FKK1227 pKa = 11.19SKK1229 pKa = 10.25GCGSASGDD1237 pKa = 4.02DD1238 pKa = 3.74TDD1240 pKa = 4.5QGDD1243 pKa = 4.04GQGCYY1248 pKa = 8.24NAKK1251 pKa = 8.79RR1252 pKa = 11.84TQQSLSLASWLASEE1266 pKa = 5.05EE1267 pKa = 4.16SLSTKK1272 pKa = 10.43EE1273 pKa = 3.74NVLIIGDD1280 pKa = 4.23LNAYY1284 pKa = 9.97AKK1286 pKa = 9.71EE1287 pKa = 4.06DD1288 pKa = 4.53PIAALEE1294 pKa = 4.22GQGFVNLVEE1303 pKa = 4.32TFQGAEE1309 pKa = 3.92AYY1311 pKa = 10.2SYY1313 pKa = 9.83TFSGEE1318 pKa = 3.73FGYY1321 pKa = 10.78LDD1323 pKa = 3.55HH1324 pKa = 7.28ALASSSLAAQAVDD1337 pKa = 3.69TIEE1340 pKa = 3.61WHH1342 pKa = 6.61INADD1346 pKa = 3.53EE1347 pKa = 4.49PFALDD1352 pKa = 3.4YY1353 pKa = 11.21NVEE1356 pKa = 4.45FKK1358 pKa = 11.08SDD1360 pKa = 3.19AQVNDD1365 pKa = 5.39FYY1367 pKa = 11.61ASDD1370 pKa = 3.66VFRR1373 pKa = 11.84VSDD1376 pKa = 4.17HH1377 pKa = 7.21DD1378 pKa = 3.72PVMMSFEE1385 pKa = 4.26LASPAVEE1392 pKa = 4.12GDD1394 pKa = 2.67IDD1396 pKa = 4.46GDD1398 pKa = 3.63MDD1400 pKa = 3.94VDD1402 pKa = 4.17YY1403 pKa = 11.75NDD1405 pKa = 3.49MRR1407 pKa = 11.84ALMNLIRR1414 pKa = 11.84AGQATLEE1421 pKa = 4.06EE1422 pKa = 4.65HH1423 pKa = 7.33DD1424 pKa = 4.49FNNDD1428 pKa = 2.68GLLNSGDD1435 pKa = 3.63VSALRR1440 pKa = 11.84SMCSRR1445 pKa = 11.84RR1446 pKa = 11.84ACSTRR1451 pKa = 3.02
Molecular weight: 154.4 kDa
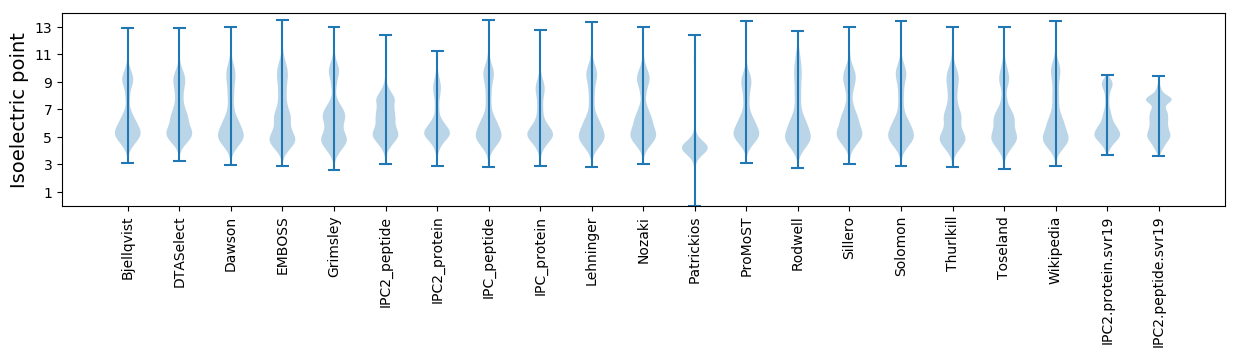
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K6WWA3|K6WWA3_9ALTE Sodium/glucose cotransporter OS=Aliiglaciecola lipolytica E3 OX=1127673 GN=sglT PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.2VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.38GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.2VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.38GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425066 |
37 |
3016 |
329.3 |
36.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.203 ± 0.034 | 0.984 ± 0.012 |
5.717 ± 0.031 | 6.121 ± 0.029 |
4.411 ± 0.026 | 6.535 ± 0.038 |
2.164 ± 0.016 | 6.64 ± 0.027 |
5.537 ± 0.038 | 10.2 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.442 ± 0.02 | 4.838 ± 0.026 |
3.828 ± 0.02 | 4.832 ± 0.034 |
4.223 ± 0.031 | 6.927 ± 0.031 |
5.277 ± 0.028 | 6.763 ± 0.03 |
1.255 ± 0.014 | 3.102 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
