
Pseudomonas phage JBD25
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
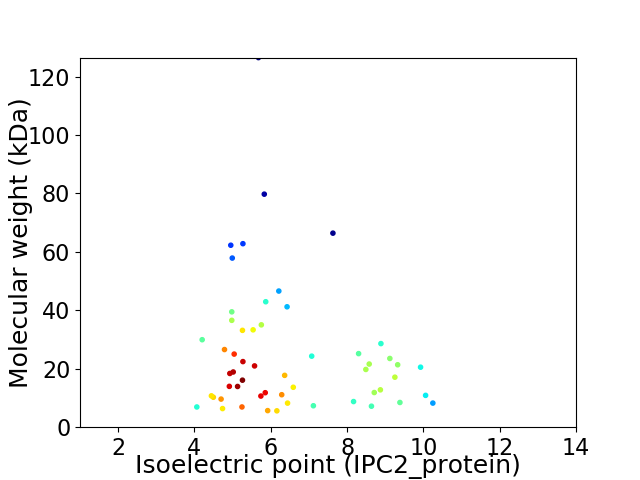
Virtual 2D-PAGE plot for 55 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9SNL4|J9SNL4_9CAUD Uncharacterized protein OS=Pseudomonas phage JBD25 OX=1225792 GN=HMPREFV_HMPID9847gp0055 PE=4 SV=1
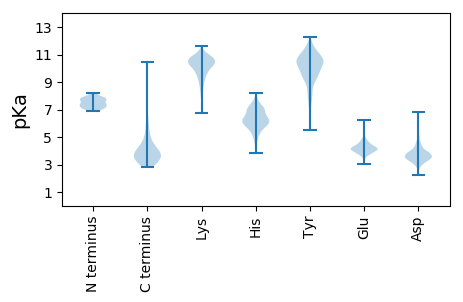
MM1 pKa = 7.46SFNSRR6 pKa = 11.84EE7 pKa = 3.92SSLVDD12 pKa = 3.44GQPVRR17 pKa = 11.84LYY19 pKa = 10.17QFSRR23 pKa = 11.84GAIRR27 pKa = 11.84WSYY30 pKa = 10.82NSSDD34 pKa = 4.7RR35 pKa = 11.84DD36 pKa = 3.1ITYY39 pKa = 10.2QNQVFRR45 pKa = 11.84TVPGGITDD53 pKa = 3.54NGIICSGDD61 pKa = 3.43PQSDD65 pKa = 3.53QFVITAPADD74 pKa = 3.6LDD76 pKa = 3.68VALLYY81 pKa = 8.52KK82 pKa = 10.05TRR84 pKa = 11.84SPSGAIDD91 pKa = 3.52LVVYY95 pKa = 10.58DD96 pKa = 3.99MHH98 pKa = 8.19YY99 pKa = 11.19GDD101 pKa = 5.63AEE103 pKa = 4.2AAVSWVGQIGDD114 pKa = 4.0VDD116 pKa = 3.86WPTVDD121 pKa = 3.06SCRR124 pKa = 11.84ITCVSEE130 pKa = 4.47DD131 pKa = 3.9EE132 pKa = 6.08LMDD135 pKa = 3.76QPGLIDD141 pKa = 3.86TYY143 pKa = 11.23CRR145 pKa = 11.84TCTAVVGDD153 pKa = 4.48HH154 pKa = 6.1RR155 pKa = 11.84CKK157 pKa = 11.04VNLVPYY163 pKa = 9.97RR164 pKa = 11.84VTLTPQSISGWVISSGVVAGYY185 pKa = 11.08ADD187 pKa = 3.46GWFTGGYY194 pKa = 8.66VEE196 pKa = 4.37WQVDD200 pKa = 3.02GDD202 pKa = 4.29NYY204 pKa = 10.6DD205 pKa = 2.73SRR207 pKa = 11.84YY208 pKa = 9.2IEE210 pKa = 4.13RR211 pKa = 11.84HH212 pKa = 5.85AGPDD216 pKa = 3.48LYY218 pKa = 10.62ILGGTEE224 pKa = 5.08GIPAGGQLRR233 pKa = 11.84VYY235 pKa = 9.42PGCDD239 pKa = 3.1GLAQTCDD246 pKa = 3.63DD247 pKa = 4.57KK248 pKa = 11.59FSNLPNFRR256 pKa = 11.84GFNAMQGKK264 pKa = 9.57SPFDD268 pKa = 4.06GDD270 pKa = 4.13QVWW273 pKa = 3.01
MM1 pKa = 7.46SFNSRR6 pKa = 11.84EE7 pKa = 3.92SSLVDD12 pKa = 3.44GQPVRR17 pKa = 11.84LYY19 pKa = 10.17QFSRR23 pKa = 11.84GAIRR27 pKa = 11.84WSYY30 pKa = 10.82NSSDD34 pKa = 4.7RR35 pKa = 11.84DD36 pKa = 3.1ITYY39 pKa = 10.2QNQVFRR45 pKa = 11.84TVPGGITDD53 pKa = 3.54NGIICSGDD61 pKa = 3.43PQSDD65 pKa = 3.53QFVITAPADD74 pKa = 3.6LDD76 pKa = 3.68VALLYY81 pKa = 8.52KK82 pKa = 10.05TRR84 pKa = 11.84SPSGAIDD91 pKa = 3.52LVVYY95 pKa = 10.58DD96 pKa = 3.99MHH98 pKa = 8.19YY99 pKa = 11.19GDD101 pKa = 5.63AEE103 pKa = 4.2AAVSWVGQIGDD114 pKa = 4.0VDD116 pKa = 3.86WPTVDD121 pKa = 3.06SCRR124 pKa = 11.84ITCVSEE130 pKa = 4.47DD131 pKa = 3.9EE132 pKa = 6.08LMDD135 pKa = 3.76QPGLIDD141 pKa = 3.86TYY143 pKa = 11.23CRR145 pKa = 11.84TCTAVVGDD153 pKa = 4.48HH154 pKa = 6.1RR155 pKa = 11.84CKK157 pKa = 11.04VNLVPYY163 pKa = 9.97RR164 pKa = 11.84VTLTPQSISGWVISSGVVAGYY185 pKa = 11.08ADD187 pKa = 3.46GWFTGGYY194 pKa = 8.66VEE196 pKa = 4.37WQVDD200 pKa = 3.02GDD202 pKa = 4.29NYY204 pKa = 10.6DD205 pKa = 2.73SRR207 pKa = 11.84YY208 pKa = 9.2IEE210 pKa = 4.13RR211 pKa = 11.84HH212 pKa = 5.85AGPDD216 pKa = 3.48LYY218 pKa = 10.62ILGGTEE224 pKa = 5.08GIPAGGQLRR233 pKa = 11.84VYY235 pKa = 9.42PGCDD239 pKa = 3.1GLAQTCDD246 pKa = 3.63DD247 pKa = 4.57KK248 pKa = 11.59FSNLPNFRR256 pKa = 11.84GFNAMQGKK264 pKa = 9.57SPFDD268 pKa = 4.06GDD270 pKa = 4.13QVWW273 pKa = 3.01
Molecular weight: 29.94 kDa
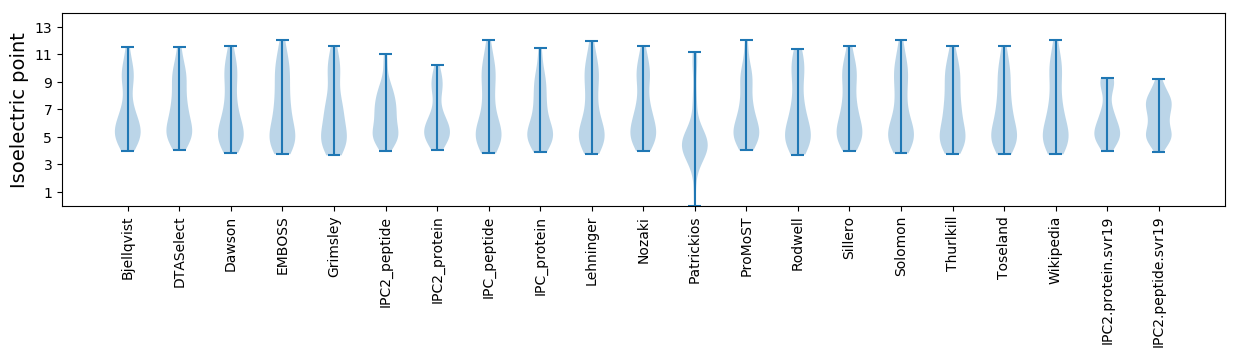
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9STP5|J9STP5_9CAUD Integrase core domain protein OS=Pseudomonas phage JBD25 OX=1225792 GN=HMPREFV_HMPID9847gp0014 PE=4 SV=1
MM1 pKa = 7.43SVEE4 pKa = 3.86AYY6 pKa = 9.26IRR8 pKa = 11.84GMAARR13 pKa = 11.84GFSRR17 pKa = 11.84SAAAAALGMHH27 pKa = 6.19WVKK30 pKa = 10.86FMDD33 pKa = 4.19LLEE36 pKa = 4.46RR37 pKa = 11.84MPDD40 pKa = 3.48IEE42 pKa = 3.84WGYY45 pKa = 9.72PYY47 pKa = 10.86KK48 pKa = 10.97SFDD51 pKa = 3.46RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.75AKK57 pKa = 8.96NLKK60 pKa = 9.27GYY62 pKa = 9.66RR63 pKa = 11.84FRR65 pKa = 11.84DD66 pKa = 3.25SEE68 pKa = 3.95GRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84SVAALRR78 pKa = 11.84AVNQARR84 pKa = 11.84RR85 pKa = 11.84HH86 pKa = 5.7EE87 pKa = 4.21YY88 pKa = 9.63TVFGVTDD95 pKa = 3.54SLSNLVKK102 pKa = 10.76RR103 pKa = 11.84FGCVAKK109 pKa = 9.97STVQKK114 pKa = 10.6RR115 pKa = 11.84LAKK118 pKa = 10.33GMSIEE123 pKa = 4.25QALTTPRR130 pKa = 11.84SDD132 pKa = 3.27HH133 pKa = 7.13LSGLKK138 pKa = 9.9RR139 pKa = 11.84KK140 pKa = 9.64PEE142 pKa = 3.72SHH144 pKa = 5.74PWKK147 pKa = 8.72RR148 pKa = 11.84AEE150 pKa = 3.75RR151 pKa = 11.84RR152 pKa = 11.84GVINHH157 pKa = 6.65RR158 pKa = 11.84EE159 pKa = 3.9RR160 pKa = 11.84QLKK163 pKa = 9.93AKK165 pKa = 9.99RR166 pKa = 11.84DD167 pKa = 3.47QRR169 pKa = 11.84QAEE172 pKa = 4.21EE173 pKa = 4.13RR174 pKa = 11.84LHH176 pKa = 6.24GG177 pKa = 4.22
MM1 pKa = 7.43SVEE4 pKa = 3.86AYY6 pKa = 9.26IRR8 pKa = 11.84GMAARR13 pKa = 11.84GFSRR17 pKa = 11.84SAAAAALGMHH27 pKa = 6.19WVKK30 pKa = 10.86FMDD33 pKa = 4.19LLEE36 pKa = 4.46RR37 pKa = 11.84MPDD40 pKa = 3.48IEE42 pKa = 3.84WGYY45 pKa = 9.72PYY47 pKa = 10.86KK48 pKa = 10.97SFDD51 pKa = 3.46RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.75AKK57 pKa = 8.96NLKK60 pKa = 9.27GYY62 pKa = 9.66RR63 pKa = 11.84FRR65 pKa = 11.84DD66 pKa = 3.25SEE68 pKa = 3.95GRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84SVAALRR78 pKa = 11.84AVNQARR84 pKa = 11.84RR85 pKa = 11.84HH86 pKa = 5.7EE87 pKa = 4.21YY88 pKa = 9.63TVFGVTDD95 pKa = 3.54SLSNLVKK102 pKa = 10.76RR103 pKa = 11.84FGCVAKK109 pKa = 9.97STVQKK114 pKa = 10.6RR115 pKa = 11.84LAKK118 pKa = 10.33GMSIEE123 pKa = 4.25QALTTPRR130 pKa = 11.84SDD132 pKa = 3.27HH133 pKa = 7.13LSGLKK138 pKa = 9.9RR139 pKa = 11.84KK140 pKa = 9.64PEE142 pKa = 3.72SHH144 pKa = 5.74PWKK147 pKa = 8.72RR148 pKa = 11.84AEE150 pKa = 3.75RR151 pKa = 11.84RR152 pKa = 11.84GVINHH157 pKa = 6.65RR158 pKa = 11.84EE159 pKa = 3.9RR160 pKa = 11.84QLKK163 pKa = 9.93AKK165 pKa = 9.99RR166 pKa = 11.84DD167 pKa = 3.47QRR169 pKa = 11.84QAEE172 pKa = 4.21EE173 pKa = 4.13RR174 pKa = 11.84LHH176 pKa = 6.24GG177 pKa = 4.22
Molecular weight: 20.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12574 |
52 |
1209 |
228.6 |
25.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.176 ± 0.703 | 0.915 ± 0.15 |
5.885 ± 0.227 | 6.259 ± 0.372 |
2.879 ± 0.177 | 7.579 ± 0.371 |
1.805 ± 0.209 | 4.143 ± 0.203 |
4.016 ± 0.321 | 10.021 ± 0.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.195 ± 0.142 | 3.046 ± 0.18 |
4.819 ± 0.328 | 5.193 ± 0.307 |
7.277 ± 0.312 | 5.694 ± 0.226 |
5.36 ± 0.323 | 6.418 ± 0.294 |
1.734 ± 0.16 | 2.585 ± 0.21 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
