
Limnothrix sp. P13C2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Pseudanabaenaceae; Limnothrix; unclassified Limnothrix
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
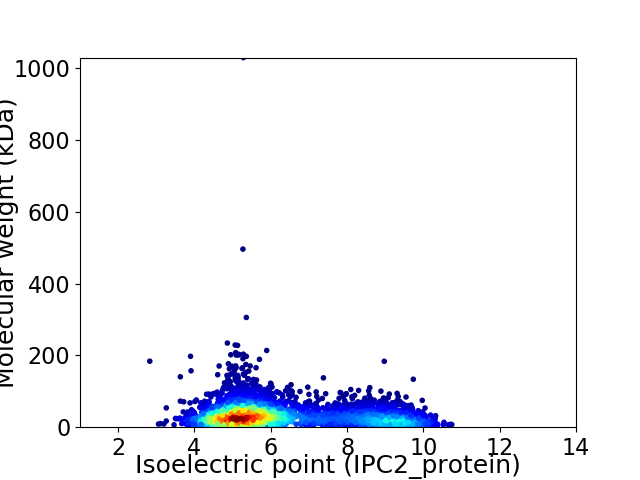
Virtual 2D-PAGE plot for 3670 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
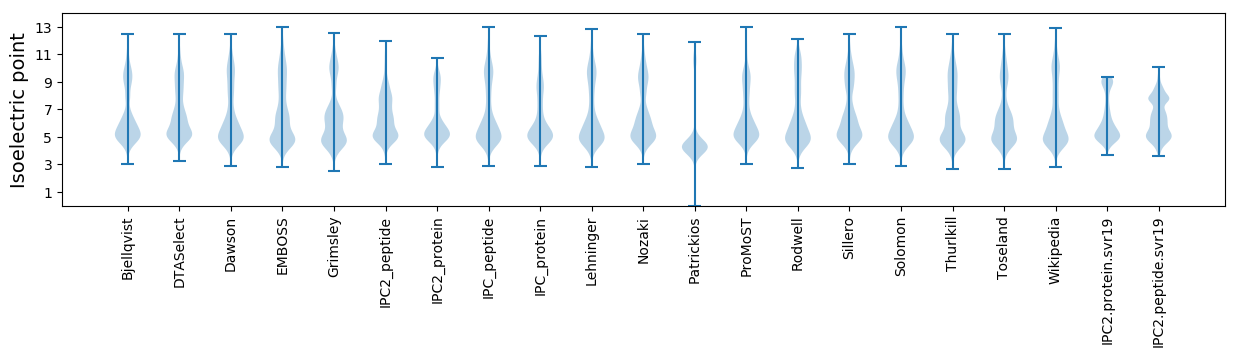
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C0V632|A0A1C0V632_9CYAN Transposase OS=Limnothrix sp. P13C2 OX=1880902 GN=BCR12_13195 PE=4 SV=1
MM1 pKa = 6.99TQFGNNLPNFLIGSGVDD18 pKa = 3.34TLIGLGDD25 pKa = 4.14ADD27 pKa = 4.19SLVSSTAGRR36 pKa = 11.84NAIFGNAGADD46 pKa = 3.56SLISQGGADD55 pKa = 4.31TIYY58 pKa = 10.39GGQGADD64 pKa = 3.34IIRR67 pKa = 11.84SRR69 pKa = 11.84GGSLLYY75 pKa = 10.56GDD77 pKa = 5.23RR78 pKa = 11.84GNDD81 pKa = 3.74TIVGEE86 pKa = 4.27ASSTGIIGGSTAGGGDD102 pKa = 3.29TVYY105 pKa = 10.86GGEE108 pKa = 4.42GDD110 pKa = 4.29DD111 pKa = 3.74SLVANSVGPSSLFGNQGNDD130 pKa = 3.05VLVAGNRR137 pKa = 11.84ADD139 pKa = 3.55TLYY142 pKa = 10.97GGQGNDD148 pKa = 3.28TLTSTVGGLLYY159 pKa = 10.7GDD161 pKa = 4.95RR162 pKa = 11.84GNDD165 pKa = 3.69SIVLSGSGVAGATTVYY181 pKa = 10.86GGDD184 pKa = 3.52GDD186 pKa = 4.75DD187 pKa = 5.22AITGGGRR194 pKa = 11.84TLLFGNQGNDD204 pKa = 3.29TITAGAADD212 pKa = 4.04SVYY215 pKa = 10.3GGSGADD221 pKa = 3.29VLTASGSGAFISGDD235 pKa = 3.45LGNDD239 pKa = 2.86ILRR242 pKa = 11.84NTGANNTLAGGEE254 pKa = 4.13GDD256 pKa = 4.07DD257 pKa = 4.79TITFDD262 pKa = 3.96GGAAFSTASGGGGKK276 pKa = 9.55NFLQILPTITGSGNVLIAGSLGDD299 pKa = 3.79TLVGAGANTIQGGAGDD315 pKa = 5.0DD316 pKa = 3.86SLVSQGSNVTLVGGAGRR333 pKa = 11.84DD334 pKa = 3.71TFNVLAGGAISGFNPGEE351 pKa = 4.14GDD353 pKa = 3.74TILSGTAPFTIIAGSTGLYY372 pKa = 8.89LTGTPNRR379 pKa = 11.84DD380 pKa = 3.37TLTGGAGNDD389 pKa = 3.57TLEE392 pKa = 4.43GLGNADD398 pKa = 3.87ILTGGAGADD407 pKa = 3.2VFLFKK412 pKa = 10.68GVAISDD418 pKa = 3.89VASVAFSNNVLQVGGTLGAGTQAAVVQDD446 pKa = 3.42VVYY449 pKa = 8.24FTSLPNPAHH458 pKa = 5.71GQSTGGNAAGTTATYY473 pKa = 10.91SFAGAGTVSSLGGTSGTFIAPPYY496 pKa = 9.58GAALSVTYY504 pKa = 8.43GTTGGVGTVVTPSFGSYY521 pKa = 8.45QATGTTAVVPVSGFGYY537 pKa = 10.46ALSGFDD543 pKa = 4.14TITDD547 pKa = 4.44FEE549 pKa = 4.83TGVDD553 pKa = 3.76QIRR556 pKa = 11.84LEE558 pKa = 4.09GRR560 pKa = 11.84LFSTGASFPNANAPGAFLSVTSITGTNVSNLVNKK594 pKa = 9.91TEE596 pKa = 4.04LLIYY600 pKa = 10.27AQDD603 pKa = 3.28SGILYY608 pKa = 9.93GRR610 pKa = 11.84AVAGGTNTNTAGTGLGSQYY629 pKa = 10.7VYY631 pKa = 11.25SPTPVPAFGTLASLNGPGDD650 pKa = 3.4QKK652 pKa = 11.53VIISSPGTVSVVGTNSNHH670 pKa = 5.75VGTVQYY676 pKa = 10.67FLLPQTPISGVAFSNSGTNTVSSVLPIPFAQVLRR710 pKa = 11.84GGAPVTGLTYY720 pKa = 10.86GRR722 pKa = 11.84DD723 pKa = 3.03IVIFF727 pKa = 4.02
MM1 pKa = 6.99TQFGNNLPNFLIGSGVDD18 pKa = 3.34TLIGLGDD25 pKa = 4.14ADD27 pKa = 4.19SLVSSTAGRR36 pKa = 11.84NAIFGNAGADD46 pKa = 3.56SLISQGGADD55 pKa = 4.31TIYY58 pKa = 10.39GGQGADD64 pKa = 3.34IIRR67 pKa = 11.84SRR69 pKa = 11.84GGSLLYY75 pKa = 10.56GDD77 pKa = 5.23RR78 pKa = 11.84GNDD81 pKa = 3.74TIVGEE86 pKa = 4.27ASSTGIIGGSTAGGGDD102 pKa = 3.29TVYY105 pKa = 10.86GGEE108 pKa = 4.42GDD110 pKa = 4.29DD111 pKa = 3.74SLVANSVGPSSLFGNQGNDD130 pKa = 3.05VLVAGNRR137 pKa = 11.84ADD139 pKa = 3.55TLYY142 pKa = 10.97GGQGNDD148 pKa = 3.28TLTSTVGGLLYY159 pKa = 10.7GDD161 pKa = 4.95RR162 pKa = 11.84GNDD165 pKa = 3.69SIVLSGSGVAGATTVYY181 pKa = 10.86GGDD184 pKa = 3.52GDD186 pKa = 4.75DD187 pKa = 5.22AITGGGRR194 pKa = 11.84TLLFGNQGNDD204 pKa = 3.29TITAGAADD212 pKa = 4.04SVYY215 pKa = 10.3GGSGADD221 pKa = 3.29VLTASGSGAFISGDD235 pKa = 3.45LGNDD239 pKa = 2.86ILRR242 pKa = 11.84NTGANNTLAGGEE254 pKa = 4.13GDD256 pKa = 4.07DD257 pKa = 4.79TITFDD262 pKa = 3.96GGAAFSTASGGGGKK276 pKa = 9.55NFLQILPTITGSGNVLIAGSLGDD299 pKa = 3.79TLVGAGANTIQGGAGDD315 pKa = 5.0DD316 pKa = 3.86SLVSQGSNVTLVGGAGRR333 pKa = 11.84DD334 pKa = 3.71TFNVLAGGAISGFNPGEE351 pKa = 4.14GDD353 pKa = 3.74TILSGTAPFTIIAGSTGLYY372 pKa = 8.89LTGTPNRR379 pKa = 11.84DD380 pKa = 3.37TLTGGAGNDD389 pKa = 3.57TLEE392 pKa = 4.43GLGNADD398 pKa = 3.87ILTGGAGADD407 pKa = 3.2VFLFKK412 pKa = 10.68GVAISDD418 pKa = 3.89VASVAFSNNVLQVGGTLGAGTQAAVVQDD446 pKa = 3.42VVYY449 pKa = 8.24FTSLPNPAHH458 pKa = 5.71GQSTGGNAAGTTATYY473 pKa = 10.91SFAGAGTVSSLGGTSGTFIAPPYY496 pKa = 9.58GAALSVTYY504 pKa = 8.43GTTGGVGTVVTPSFGSYY521 pKa = 8.45QATGTTAVVPVSGFGYY537 pKa = 10.46ALSGFDD543 pKa = 4.14TITDD547 pKa = 4.44FEE549 pKa = 4.83TGVDD553 pKa = 3.76QIRR556 pKa = 11.84LEE558 pKa = 4.09GRR560 pKa = 11.84LFSTGASFPNANAPGAFLSVTSITGTNVSNLVNKK594 pKa = 9.91TEE596 pKa = 4.04LLIYY600 pKa = 10.27AQDD603 pKa = 3.28SGILYY608 pKa = 9.93GRR610 pKa = 11.84AVAGGTNTNTAGTGLGSQYY629 pKa = 10.7VYY631 pKa = 11.25SPTPVPAFGTLASLNGPGDD650 pKa = 3.4QKK652 pKa = 11.53VIISSPGTVSVVGTNSNHH670 pKa = 5.75VGTVQYY676 pKa = 10.67FLLPQTPISGVAFSNSGTNTVSSVLPIPFAQVLRR710 pKa = 11.84GGAPVTGLTYY720 pKa = 10.86GRR722 pKa = 11.84DD723 pKa = 3.03IVIFF727 pKa = 4.02
Molecular weight: 70.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C0V266|A0A1C0V266_9CYAN XisI protein OS=Limnothrix sp. P13C2 OX=1880902 GN=BCR12_01825 PE=4 SV=1
MM1 pKa = 7.34AKK3 pKa = 9.55SWVSRR8 pKa = 11.84SIEE11 pKa = 4.05ALSDD15 pKa = 3.89LVWQRR20 pKa = 11.84SCEE23 pKa = 4.21LCDD26 pKa = 4.76RR27 pKa = 11.84PAKK30 pKa = 10.37HH31 pKa = 6.34QGLCLACDD39 pKa = 3.53RR40 pKa = 11.84RR41 pKa = 11.84LLASQLPLEE50 pKa = 4.44QRR52 pKa = 11.84WGRR55 pKa = 11.84LPSALVSSAPLFLAWGLYY73 pKa = 10.21DD74 pKa = 4.12GSLKK78 pKa = 10.49RR79 pKa = 11.84AIAALKK85 pKa = 10.71YY86 pKa = 9.78EE87 pKa = 4.26GHH89 pKa = 7.08PKK91 pKa = 9.93LAATLGQGLGRR102 pKa = 11.84LWLEE106 pKa = 4.32PGTEE110 pKa = 3.79RR111 pKa = 11.84AMATPWQQLDD121 pKa = 3.62RR122 pKa = 11.84AAPWVIPIPLHH133 pKa = 5.12EE134 pKa = 5.28ARR136 pKa = 11.84LQKK139 pKa = 10.55RR140 pKa = 11.84GFNQAEE146 pKa = 4.04LLAKK150 pKa = 9.63HH151 pKa = 5.89FCDD154 pKa = 3.33VTGYY158 pKa = 11.02RR159 pKa = 11.84LIPNGLQRR167 pKa = 11.84IRR169 pKa = 11.84DD170 pKa = 3.82TQAQFGLGQGDD181 pKa = 3.29RR182 pKa = 11.84AANVQGAFALGPGLGQQRR200 pKa = 11.84PRR202 pKa = 11.84SVILLDD208 pKa = 5.2DD209 pKa = 4.8IYY211 pKa = 11.13TSGATVQAASNSLRR225 pKa = 11.84AGGWTVAAVVVVARR239 pKa = 11.84SAQRR243 pKa = 11.84PAPGTHH249 pKa = 6.92PPTRR253 pKa = 11.84SAPTRR258 pKa = 11.84PKK260 pKa = 10.68RR261 pKa = 11.84RR262 pKa = 11.84DD263 pKa = 3.1SS264 pKa = 3.46
MM1 pKa = 7.34AKK3 pKa = 9.55SWVSRR8 pKa = 11.84SIEE11 pKa = 4.05ALSDD15 pKa = 3.89LVWQRR20 pKa = 11.84SCEE23 pKa = 4.21LCDD26 pKa = 4.76RR27 pKa = 11.84PAKK30 pKa = 10.37HH31 pKa = 6.34QGLCLACDD39 pKa = 3.53RR40 pKa = 11.84RR41 pKa = 11.84LLASQLPLEE50 pKa = 4.44QRR52 pKa = 11.84WGRR55 pKa = 11.84LPSALVSSAPLFLAWGLYY73 pKa = 10.21DD74 pKa = 4.12GSLKK78 pKa = 10.49RR79 pKa = 11.84AIAALKK85 pKa = 10.71YY86 pKa = 9.78EE87 pKa = 4.26GHH89 pKa = 7.08PKK91 pKa = 9.93LAATLGQGLGRR102 pKa = 11.84LWLEE106 pKa = 4.32PGTEE110 pKa = 3.79RR111 pKa = 11.84AMATPWQQLDD121 pKa = 3.62RR122 pKa = 11.84AAPWVIPIPLHH133 pKa = 5.12EE134 pKa = 5.28ARR136 pKa = 11.84LQKK139 pKa = 10.55RR140 pKa = 11.84GFNQAEE146 pKa = 4.04LLAKK150 pKa = 9.63HH151 pKa = 5.89FCDD154 pKa = 3.33VTGYY158 pKa = 11.02RR159 pKa = 11.84LIPNGLQRR167 pKa = 11.84IRR169 pKa = 11.84DD170 pKa = 3.82TQAQFGLGQGDD181 pKa = 3.29RR182 pKa = 11.84AANVQGAFALGPGLGQQRR200 pKa = 11.84PRR202 pKa = 11.84SVILLDD208 pKa = 5.2DD209 pKa = 4.8IYY211 pKa = 11.13TSGATVQAASNSLRR225 pKa = 11.84AGGWTVAAVVVVARR239 pKa = 11.84SAQRR243 pKa = 11.84PAPGTHH249 pKa = 6.92PPTRR253 pKa = 11.84SAPTRR258 pKa = 11.84PKK260 pKa = 10.68RR261 pKa = 11.84RR262 pKa = 11.84DD263 pKa = 3.1SS264 pKa = 3.46
Molecular weight: 28.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208963 |
29 |
9652 |
329.4 |
36.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.418 ± 0.058 | 0.97 ± 0.012 |
5.526 ± 0.037 | 5.643 ± 0.041 |
3.479 ± 0.026 | 7.461 ± 0.047 |
1.794 ± 0.018 | 5.348 ± 0.032 |
2.955 ± 0.037 | 11.479 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.821 ± 0.018 | 3.171 ± 0.03 |
5.733 ± 0.045 | 5.363 ± 0.034 |
6.693 ± 0.036 | 5.983 ± 0.033 |
5.306 ± 0.026 | 6.616 ± 0.031 |
1.752 ± 0.024 | 2.487 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
