
Armillaria solidipes
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Physalacriaceae; Armillaria
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
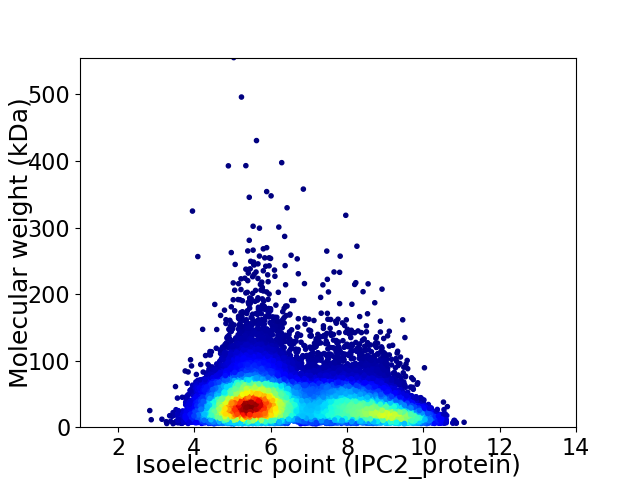
Virtual 2D-PAGE plot for 20715 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H3CLT8|A0A2H3CLT8_9AGAR FAS1 domain-containing protein OS=Armillaria solidipes OX=1076256 GN=ARMSODRAFT_1010383 PE=4 SV=1
MM1 pKa = 6.9KK2 pKa = 9.87TLVLSLLSIPFSAVAQYY19 pKa = 7.79TWKK22 pKa = 10.31NVKK25 pKa = 9.87IGGGGGFVPGIVFNPSEE42 pKa = 4.14EE43 pKa = 3.74GLAYY47 pKa = 10.6VRR49 pKa = 11.84TDD51 pKa = 2.33IGGAYY56 pKa = 9.96RR57 pKa = 11.84LNDD60 pKa = 4.05DD61 pKa = 4.65DD62 pKa = 4.02TWTPLTDD69 pKa = 3.14WVDD72 pKa = 3.27NDD74 pKa = 3.56RR75 pKa = 11.84WNMWGVDD82 pKa = 3.84ALATDD87 pKa = 3.79PVEE90 pKa = 4.25TNRR93 pKa = 11.84LYY95 pKa = 11.04LATGMYY101 pKa = 8.35TNSWDD106 pKa = 3.67PDD108 pKa = 3.38NGHH111 pKa = 6.67ILVSEE116 pKa = 4.68DD117 pKa = 3.36YY118 pKa = 11.0GEE120 pKa = 4.96SFAQVDD126 pKa = 4.29LPFKK130 pKa = 10.91VGGNMPGRR138 pKa = 11.84GVGEE142 pKa = 4.12RR143 pKa = 11.84LAVDD147 pKa = 3.76PNSNNILFFGARR159 pKa = 11.84SGNGLYY165 pKa = 10.58KK166 pKa = 10.06STDD169 pKa = 3.5YY170 pKa = 11.45GSTWTQVSSLPDD182 pKa = 3.1VGTYY186 pKa = 10.28VVDD189 pKa = 3.6ATDD192 pKa = 3.34TSGYY196 pKa = 10.72NSDD199 pKa = 3.8PLGIAFVTFDD209 pKa = 3.91STSGSSGSPTPRR221 pKa = 11.84VFVGVASNGTDD232 pKa = 3.1NIFVSEE238 pKa = 4.33DD239 pKa = 3.65AGDD242 pKa = 3.32TWTAIAGQQQTYY254 pKa = 9.21FPHH257 pKa = 6.91KK258 pKa = 10.37GVLSPDD264 pKa = 3.08EE265 pKa = 4.2GLLYY269 pKa = 10.7VSYY272 pKa = 11.46SDD274 pKa = 3.5GTGPYY279 pKa = 10.01DD280 pKa = 3.46GTLGAVYY287 pKa = 10.0KK288 pKa = 10.7YY289 pKa = 10.77DD290 pKa = 3.38IANATWTDD298 pKa = 3.2ITPVSGSDD306 pKa = 3.15LSFGFGGLSVDD317 pKa = 3.79VKK319 pKa = 11.41NPGTLMVAALNLWWPDD335 pKa = 3.16GQIYY339 pKa = 10.44RR340 pKa = 11.84STDD343 pKa = 2.97SGATWSTFWTWGAYY357 pKa = 9.26PEE359 pKa = 4.07INKK362 pKa = 10.02YY363 pKa = 10.5YY364 pKa = 10.5SYY366 pKa = 11.31SDD368 pKa = 4.29SLAPWLGVDD377 pKa = 4.74YY378 pKa = 11.46VDD380 pKa = 3.63TTLGDD385 pKa = 3.8KK386 pKa = 10.73QIGWMMEE393 pKa = 3.94ALSIDD398 pKa = 4.37PFDD401 pKa = 4.74SDD403 pKa = 3.46HH404 pKa = 6.59FLYY407 pKa = 9.0GTGATIYY414 pKa = 10.49GSRR417 pKa = 11.84DD418 pKa = 2.97LTLWDD423 pKa = 3.65SVHH426 pKa = 6.48NISLSSMADD435 pKa = 3.66GIEE438 pKa = 4.06EE439 pKa = 4.12TAVQALISPPGGPSLISGVSDD460 pKa = 2.82IQGFVHH466 pKa = 6.83TDD468 pKa = 3.07LGTPTTSFSSPVWTSVVDD486 pKa = 4.17LDD488 pKa = 4.17YY489 pKa = 11.57AGNVPTNIIRR499 pKa = 11.84LGNGDD504 pKa = 3.83SSTGKK509 pKa = 9.78QVAISTDD516 pKa = 3.44SGTTWSQDD524 pKa = 2.25TGAPDD529 pKa = 3.4NVSGGKK535 pKa = 9.21IAISADD541 pKa = 2.9GDD543 pKa = 4.0TVLWRR548 pKa = 11.84TSSNGVLVSQNTSAFTTVSTLPTSAVIASDD578 pKa = 3.23KK579 pKa = 10.01TNNSVFYY586 pKa = 9.69GASSSSFYY594 pKa = 11.03VSNDD598 pKa = 3.07TGSTFSAVSTLGNSTSPVKK617 pKa = 10.43IVVNPDD623 pKa = 2.7VAGDD627 pKa = 3.5VWVSTDD633 pKa = 2.7VGLFHH638 pKa = 6.53STDD641 pKa = 3.43YY642 pKa = 11.38GSTFMAIEE650 pKa = 4.4SVSQAWAIALGTAATDD666 pKa = 3.46GGYY669 pKa = 9.48PAVFAAANIGDD680 pKa = 3.65GVGYY684 pKa = 10.27YY685 pKa = 10.44RR686 pKa = 11.84SDD688 pKa = 3.87DD689 pKa = 3.96EE690 pKa = 4.68GATWTQINDD699 pKa = 3.19SSLGFSSVSANVMAADD715 pKa = 3.41PRR717 pKa = 11.84VYY719 pKa = 10.28GRR721 pKa = 11.84VYY723 pKa = 10.17IGTNGRR729 pKa = 11.84GIFYY733 pKa = 10.77GDD735 pKa = 3.25ISSSGTSTSSTVGPSSSAAVSTSAAAASSAVTSSIQASPSATTASSSEE783 pKa = 4.19NINGSGSQRR792 pKa = 11.84LL793 pKa = 3.52
MM1 pKa = 6.9KK2 pKa = 9.87TLVLSLLSIPFSAVAQYY19 pKa = 7.79TWKK22 pKa = 10.31NVKK25 pKa = 9.87IGGGGGFVPGIVFNPSEE42 pKa = 4.14EE43 pKa = 3.74GLAYY47 pKa = 10.6VRR49 pKa = 11.84TDD51 pKa = 2.33IGGAYY56 pKa = 9.96RR57 pKa = 11.84LNDD60 pKa = 4.05DD61 pKa = 4.65DD62 pKa = 4.02TWTPLTDD69 pKa = 3.14WVDD72 pKa = 3.27NDD74 pKa = 3.56RR75 pKa = 11.84WNMWGVDD82 pKa = 3.84ALATDD87 pKa = 3.79PVEE90 pKa = 4.25TNRR93 pKa = 11.84LYY95 pKa = 11.04LATGMYY101 pKa = 8.35TNSWDD106 pKa = 3.67PDD108 pKa = 3.38NGHH111 pKa = 6.67ILVSEE116 pKa = 4.68DD117 pKa = 3.36YY118 pKa = 11.0GEE120 pKa = 4.96SFAQVDD126 pKa = 4.29LPFKK130 pKa = 10.91VGGNMPGRR138 pKa = 11.84GVGEE142 pKa = 4.12RR143 pKa = 11.84LAVDD147 pKa = 3.76PNSNNILFFGARR159 pKa = 11.84SGNGLYY165 pKa = 10.58KK166 pKa = 10.06STDD169 pKa = 3.5YY170 pKa = 11.45GSTWTQVSSLPDD182 pKa = 3.1VGTYY186 pKa = 10.28VVDD189 pKa = 3.6ATDD192 pKa = 3.34TSGYY196 pKa = 10.72NSDD199 pKa = 3.8PLGIAFVTFDD209 pKa = 3.91STSGSSGSPTPRR221 pKa = 11.84VFVGVASNGTDD232 pKa = 3.1NIFVSEE238 pKa = 4.33DD239 pKa = 3.65AGDD242 pKa = 3.32TWTAIAGQQQTYY254 pKa = 9.21FPHH257 pKa = 6.91KK258 pKa = 10.37GVLSPDD264 pKa = 3.08EE265 pKa = 4.2GLLYY269 pKa = 10.7VSYY272 pKa = 11.46SDD274 pKa = 3.5GTGPYY279 pKa = 10.01DD280 pKa = 3.46GTLGAVYY287 pKa = 10.0KK288 pKa = 10.7YY289 pKa = 10.77DD290 pKa = 3.38IANATWTDD298 pKa = 3.2ITPVSGSDD306 pKa = 3.15LSFGFGGLSVDD317 pKa = 3.79VKK319 pKa = 11.41NPGTLMVAALNLWWPDD335 pKa = 3.16GQIYY339 pKa = 10.44RR340 pKa = 11.84STDD343 pKa = 2.97SGATWSTFWTWGAYY357 pKa = 9.26PEE359 pKa = 4.07INKK362 pKa = 10.02YY363 pKa = 10.5YY364 pKa = 10.5SYY366 pKa = 11.31SDD368 pKa = 4.29SLAPWLGVDD377 pKa = 4.74YY378 pKa = 11.46VDD380 pKa = 3.63TTLGDD385 pKa = 3.8KK386 pKa = 10.73QIGWMMEE393 pKa = 3.94ALSIDD398 pKa = 4.37PFDD401 pKa = 4.74SDD403 pKa = 3.46HH404 pKa = 6.59FLYY407 pKa = 9.0GTGATIYY414 pKa = 10.49GSRR417 pKa = 11.84DD418 pKa = 2.97LTLWDD423 pKa = 3.65SVHH426 pKa = 6.48NISLSSMADD435 pKa = 3.66GIEE438 pKa = 4.06EE439 pKa = 4.12TAVQALISPPGGPSLISGVSDD460 pKa = 2.82IQGFVHH466 pKa = 6.83TDD468 pKa = 3.07LGTPTTSFSSPVWTSVVDD486 pKa = 4.17LDD488 pKa = 4.17YY489 pKa = 11.57AGNVPTNIIRR499 pKa = 11.84LGNGDD504 pKa = 3.83SSTGKK509 pKa = 9.78QVAISTDD516 pKa = 3.44SGTTWSQDD524 pKa = 2.25TGAPDD529 pKa = 3.4NVSGGKK535 pKa = 9.21IAISADD541 pKa = 2.9GDD543 pKa = 4.0TVLWRR548 pKa = 11.84TSSNGVLVSQNTSAFTTVSTLPTSAVIASDD578 pKa = 3.23KK579 pKa = 10.01TNNSVFYY586 pKa = 9.69GASSSSFYY594 pKa = 11.03VSNDD598 pKa = 3.07TGSTFSAVSTLGNSTSPVKK617 pKa = 10.43IVVNPDD623 pKa = 2.7VAGDD627 pKa = 3.5VWVSTDD633 pKa = 2.7VGLFHH638 pKa = 6.53STDD641 pKa = 3.43YY642 pKa = 11.38GSTFMAIEE650 pKa = 4.4SVSQAWAIALGTAATDD666 pKa = 3.46GGYY669 pKa = 9.48PAVFAAANIGDD680 pKa = 3.65GVGYY684 pKa = 10.27YY685 pKa = 10.44RR686 pKa = 11.84SDD688 pKa = 3.87DD689 pKa = 3.96EE690 pKa = 4.68GATWTQINDD699 pKa = 3.19SSLGFSSVSANVMAADD715 pKa = 3.41PRR717 pKa = 11.84VYY719 pKa = 10.28GRR721 pKa = 11.84VYY723 pKa = 10.17IGTNGRR729 pKa = 11.84GIFYY733 pKa = 10.77GDD735 pKa = 3.25ISSSGTSTSSTVGPSSSAAVSTSAAAASSAVTSSIQASPSATTASSSEE783 pKa = 4.19NINGSGSQRR792 pKa = 11.84LL793 pKa = 3.52
Molecular weight: 83.0 kDa
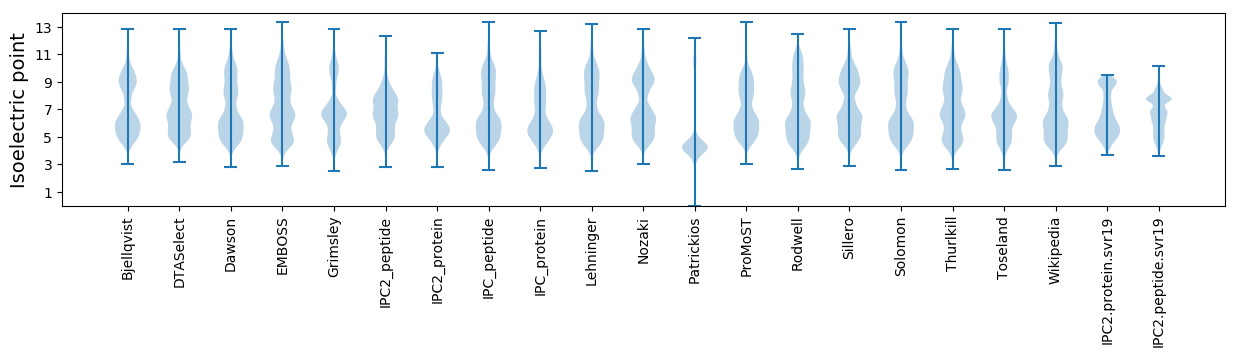
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H3C3P5|A0A2H3C3P5_9AGAR Uncharacterized protein OS=Armillaria solidipes OX=1076256 GN=ARMSODRAFT_1017547 PE=4 SV=1
SS1 pKa = 5.73TQVQRR6 pKa = 11.84PASAIQRR13 pKa = 11.84PTSIIQRR20 pKa = 11.84PASVVQRR27 pKa = 11.84PSSVIQRR34 pKa = 11.84PASITQRR41 pKa = 11.84PASAAQRR48 pKa = 11.84STLAIQHH55 pKa = 6.54PASAAQRR62 pKa = 11.84QPLAVQQQ69 pKa = 3.7
SS1 pKa = 5.73TQVQRR6 pKa = 11.84PASAIQRR13 pKa = 11.84PTSIIQRR20 pKa = 11.84PASVVQRR27 pKa = 11.84PSSVIQRR34 pKa = 11.84PASITQRR41 pKa = 11.84PASAAQRR48 pKa = 11.84STLAIQHH55 pKa = 6.54PASAAQRR62 pKa = 11.84QPLAVQQQ69 pKa = 3.7
Molecular weight: 7.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7685379 |
49 |
4986 |
371.0 |
41.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.774 ± 0.017 | 1.494 ± 0.007 |
5.687 ± 0.012 | 5.675 ± 0.016 |
3.952 ± 0.011 | 6.192 ± 0.017 |
2.593 ± 0.008 | 5.295 ± 0.011 |
4.444 ± 0.018 | 9.528 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.206 ± 0.007 | 3.473 ± 0.009 |
6.162 ± 0.018 | 3.554 ± 0.01 |
6.103 ± 0.015 | 8.803 ± 0.022 |
6.086 ± 0.011 | 6.501 ± 0.011 |
1.579 ± 0.007 | 2.899 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
