
Verticillium dahliae partitivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
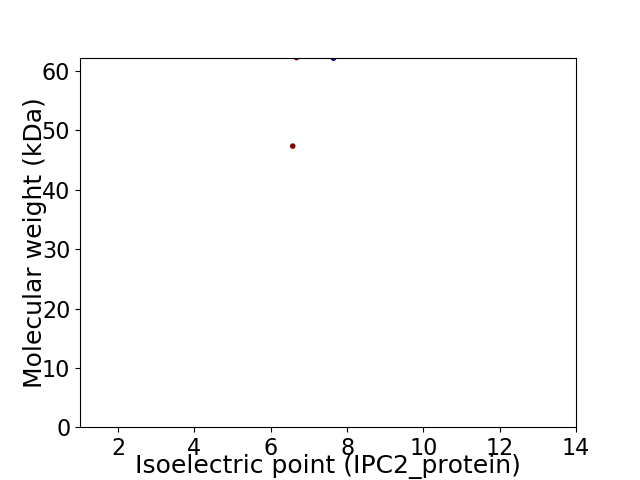
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M3LA90|A0A0M3LA90_9VIRU RNA-dependent RNA polymerase OS=Verticillium dahliae partitivirus 1 OX=1310424 GN=RdRp PE=4 SV=1
MM1 pKa = 8.04ADD3 pKa = 3.53TQSQLPSTVAPSDD16 pKa = 3.72SASTAGRR23 pKa = 11.84KK24 pKa = 8.95SKK26 pKa = 10.2PGKK29 pKa = 10.18AEE31 pKa = 3.65RR32 pKa = 11.84QARR35 pKa = 11.84RR36 pKa = 11.84SAVGSQPGQAASASKK51 pKa = 10.84ASAFSSGISAPKK63 pKa = 8.53PQPGKK68 pKa = 10.6YY69 pKa = 9.01PVVFQTGAGEE79 pKa = 4.02PARR82 pKa = 11.84DD83 pKa = 3.25QTFALDD89 pKa = 3.5EE90 pKa = 4.38RR91 pKa = 11.84VLEE94 pKa = 4.24RR95 pKa = 11.84SLRR98 pKa = 11.84TFPDD102 pKa = 2.92RR103 pKa = 11.84FTFNAKK109 pKa = 8.79YY110 pKa = 10.89AEE112 pKa = 4.41FKK114 pKa = 10.66SHH116 pKa = 7.52AEE118 pKa = 3.79IDD120 pKa = 3.56DD121 pKa = 4.0HH122 pKa = 7.05EE123 pKa = 4.45FTKK126 pKa = 10.99DD127 pKa = 3.37LVVAALLRR135 pKa = 11.84LAQQVVHH142 pKa = 5.67SHH144 pKa = 5.41VNMGLPQGDD153 pKa = 4.28FAPVASSDD161 pKa = 3.46VRR163 pKa = 11.84VPASLAAFISQFGEE177 pKa = 4.1FSVPTLGTRR186 pKa = 11.84FLFSDD191 pKa = 3.82YY192 pKa = 10.61EE193 pKa = 4.14NTVRR197 pKa = 11.84SIVWMAEE204 pKa = 3.74GVKK207 pKa = 10.56RR208 pKa = 11.84NGIGGAPLKK217 pKa = 10.64RR218 pKa = 11.84SWLPVRR224 pKa = 11.84PSDD227 pKa = 3.33GHH229 pKa = 5.6TKK231 pKa = 9.31TVIASRR237 pKa = 11.84LCEE240 pKa = 3.97LLAQAEE246 pKa = 4.37LAVDD250 pKa = 4.04PDD252 pKa = 3.8ALEE255 pKa = 4.59GGVLSGEE262 pKa = 4.45IPDD265 pKa = 3.29AWEE268 pKa = 4.08SIKK271 pKa = 10.71PVLGDD276 pKa = 3.09NDD278 pKa = 3.4EE279 pKa = 4.21RR280 pKa = 11.84RR281 pKa = 11.84DD282 pKa = 3.69RR283 pKa = 11.84FDD285 pKa = 5.13FLFKK289 pKa = 10.45SYY291 pKa = 10.57RR292 pKa = 11.84DD293 pKa = 3.35APAFVTAFTTASASGVLAEE312 pKa = 6.06LDD314 pKa = 4.06LSWDD318 pKa = 3.78RR319 pKa = 11.84PSAGHH324 pKa = 6.45VDD326 pKa = 2.89WTFNPKK332 pKa = 9.7EE333 pKa = 4.09VFTRR337 pKa = 11.84LSDD340 pKa = 2.85AWARR344 pKa = 11.84RR345 pKa = 11.84STTYY349 pKa = 11.61ALFFEE354 pKa = 5.28LSSSQMTRR362 pKa = 11.84NVSAGSQSQMARR374 pKa = 11.84VNTRR378 pKa = 11.84DD379 pKa = 2.96GVTVVKK385 pKa = 9.06THH387 pKa = 6.95LGLSAPEE394 pKa = 4.12FSLVACFPASVIFTGGLTRR413 pKa = 11.84RR414 pKa = 11.84VVVTTPLSVEE424 pKa = 3.73QRR426 pKa = 11.84ATEE429 pKa = 4.28FVQMDD434 pKa = 3.21WRR436 pKa = 5.26
MM1 pKa = 8.04ADD3 pKa = 3.53TQSQLPSTVAPSDD16 pKa = 3.72SASTAGRR23 pKa = 11.84KK24 pKa = 8.95SKK26 pKa = 10.2PGKK29 pKa = 10.18AEE31 pKa = 3.65RR32 pKa = 11.84QARR35 pKa = 11.84RR36 pKa = 11.84SAVGSQPGQAASASKK51 pKa = 10.84ASAFSSGISAPKK63 pKa = 8.53PQPGKK68 pKa = 10.6YY69 pKa = 9.01PVVFQTGAGEE79 pKa = 4.02PARR82 pKa = 11.84DD83 pKa = 3.25QTFALDD89 pKa = 3.5EE90 pKa = 4.38RR91 pKa = 11.84VLEE94 pKa = 4.24RR95 pKa = 11.84SLRR98 pKa = 11.84TFPDD102 pKa = 2.92RR103 pKa = 11.84FTFNAKK109 pKa = 8.79YY110 pKa = 10.89AEE112 pKa = 4.41FKK114 pKa = 10.66SHH116 pKa = 7.52AEE118 pKa = 3.79IDD120 pKa = 3.56DD121 pKa = 4.0HH122 pKa = 7.05EE123 pKa = 4.45FTKK126 pKa = 10.99DD127 pKa = 3.37LVVAALLRR135 pKa = 11.84LAQQVVHH142 pKa = 5.67SHH144 pKa = 5.41VNMGLPQGDD153 pKa = 4.28FAPVASSDD161 pKa = 3.46VRR163 pKa = 11.84VPASLAAFISQFGEE177 pKa = 4.1FSVPTLGTRR186 pKa = 11.84FLFSDD191 pKa = 3.82YY192 pKa = 10.61EE193 pKa = 4.14NTVRR197 pKa = 11.84SIVWMAEE204 pKa = 3.74GVKK207 pKa = 10.56RR208 pKa = 11.84NGIGGAPLKK217 pKa = 10.64RR218 pKa = 11.84SWLPVRR224 pKa = 11.84PSDD227 pKa = 3.33GHH229 pKa = 5.6TKK231 pKa = 9.31TVIASRR237 pKa = 11.84LCEE240 pKa = 3.97LLAQAEE246 pKa = 4.37LAVDD250 pKa = 4.04PDD252 pKa = 3.8ALEE255 pKa = 4.59GGVLSGEE262 pKa = 4.45IPDD265 pKa = 3.29AWEE268 pKa = 4.08SIKK271 pKa = 10.71PVLGDD276 pKa = 3.09NDD278 pKa = 3.4EE279 pKa = 4.21RR280 pKa = 11.84RR281 pKa = 11.84DD282 pKa = 3.69RR283 pKa = 11.84FDD285 pKa = 5.13FLFKK289 pKa = 10.45SYY291 pKa = 10.57RR292 pKa = 11.84DD293 pKa = 3.35APAFVTAFTTASASGVLAEE312 pKa = 6.06LDD314 pKa = 4.06LSWDD318 pKa = 3.78RR319 pKa = 11.84PSAGHH324 pKa = 6.45VDD326 pKa = 2.89WTFNPKK332 pKa = 9.7EE333 pKa = 4.09VFTRR337 pKa = 11.84LSDD340 pKa = 2.85AWARR344 pKa = 11.84RR345 pKa = 11.84STTYY349 pKa = 11.61ALFFEE354 pKa = 5.28LSSSQMTRR362 pKa = 11.84NVSAGSQSQMARR374 pKa = 11.84VNTRR378 pKa = 11.84DD379 pKa = 2.96GVTVVKK385 pKa = 9.06THH387 pKa = 6.95LGLSAPEE394 pKa = 4.12FSLVACFPASVIFTGGLTRR413 pKa = 11.84RR414 pKa = 11.84VVVTTPLSVEE424 pKa = 3.73QRR426 pKa = 11.84ATEE429 pKa = 4.28FVQMDD434 pKa = 3.21WRR436 pKa = 5.26
Molecular weight: 47.37 kDa
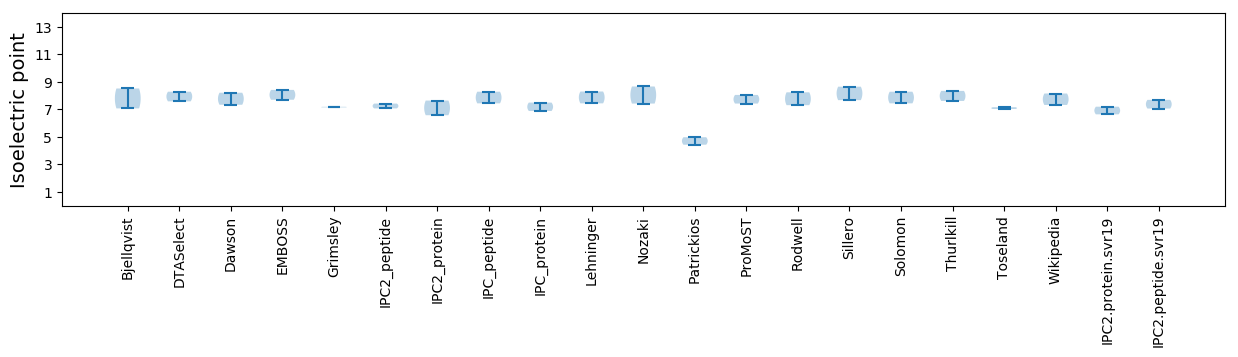
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M3LA90|A0A0M3LA90_9VIRU RNA-dependent RNA polymerase OS=Verticillium dahliae partitivirus 1 OX=1310424 GN=RdRp PE=4 SV=1
MM1 pKa = 7.93EE2 pKa = 6.77DD3 pKa = 3.42FTQDD7 pKa = 3.31PTTHH11 pKa = 6.67NIVAEE16 pKa = 4.58GSHH19 pKa = 7.06LIDD22 pKa = 4.56ALHH25 pKa = 6.35LRR27 pKa = 11.84PPKK30 pKa = 10.28SRR32 pKa = 11.84SATSEE37 pKa = 4.03DD38 pKa = 3.8VVSSNFRR45 pKa = 11.84SPNLAEE51 pKa = 3.65IAKK54 pKa = 9.21YY55 pKa = 10.3GGYY58 pKa = 7.94STYY61 pKa = 11.11SSNSNTDD68 pKa = 2.97PYY70 pKa = 11.29VRR72 pKa = 11.84EE73 pKa = 3.98TLKK76 pKa = 10.98LFSRR80 pKa = 11.84DD81 pKa = 3.06TYY83 pKa = 10.88EE84 pKa = 5.05DD85 pKa = 2.74IRR87 pKa = 11.84GFTRR91 pKa = 11.84RR92 pKa = 11.84PQGTPGMYY100 pKa = 8.42TALKK104 pKa = 9.81KK105 pKa = 10.83FSGEE109 pKa = 3.94RR110 pKa = 11.84NTFGDD115 pKa = 4.73LSPSQQSSMRR125 pKa = 11.84RR126 pKa = 11.84AIGKK130 pKa = 9.39AKK132 pKa = 10.31KK133 pKa = 10.09AFKK136 pKa = 10.39LPYY139 pKa = 9.55KK140 pKa = 10.34RR141 pKa = 11.84EE142 pKa = 3.91PLDD145 pKa = 3.15WHH147 pKa = 6.48EE148 pKa = 4.21VGQFLRR154 pKa = 11.84RR155 pKa = 11.84DD156 pKa = 3.56TAAGATFMGQKK167 pKa = 10.11KK168 pKa = 9.68GDD170 pKa = 3.59VMEE173 pKa = 5.62AIYY176 pKa = 10.8HH177 pKa = 5.12EE178 pKa = 4.58ARR180 pKa = 11.84WLGHH184 pKa = 6.1RR185 pKa = 11.84MKK187 pKa = 10.64QDD189 pKa = 2.56GRR191 pKa = 11.84AGFDD195 pKa = 3.4PTRR198 pKa = 11.84MRR200 pKa = 11.84FPPCLAGQRR209 pKa = 11.84GGMSEE214 pKa = 3.87IDD216 pKa = 3.64DD217 pKa = 4.28PKK219 pKa = 11.01TRR221 pKa = 11.84LVWIYY226 pKa = 8.84PAEE229 pKa = 4.02MLVVEE234 pKa = 5.03GFYY237 pKa = 11.12APLMYY242 pKa = 9.66RR243 pKa = 11.84DD244 pKa = 4.75FMSDD248 pKa = 3.17PNSPMLNGKK257 pKa = 8.31SAQRR261 pKa = 11.84LYY263 pKa = 10.74TEE265 pKa = 4.78WCCKK269 pKa = 10.29LRR271 pKa = 11.84DD272 pKa = 3.54GEE274 pKa = 4.38TLYY277 pKa = 11.52GIDD280 pKa = 4.38FSSFDD285 pKa = 3.59TKK287 pKa = 10.85VPAWLIRR294 pKa = 11.84VAFDD298 pKa = 4.45ILRR301 pKa = 11.84QNINFEE307 pKa = 4.4TFGGKK312 pKa = 9.0PVDD315 pKa = 3.87KK316 pKa = 10.81QDD318 pKa = 3.18AQKK321 pKa = 9.9WRR323 pKa = 11.84NVWDD327 pKa = 3.45AMVWYY332 pKa = 8.66FINTPILMPDD342 pKa = 2.44GRR344 pKa = 11.84MFRR347 pKa = 11.84KK348 pKa = 9.65FRR350 pKa = 11.84GVPSGSWWTQMIDD363 pKa = 3.35SVVNHH368 pKa = 6.47ILIDD372 pKa = 3.82YY373 pKa = 9.97LADD376 pKa = 3.6CQRR379 pKa = 11.84VEE381 pKa = 3.79IRR383 pKa = 11.84NLRR386 pKa = 11.84VLGDD390 pKa = 3.83DD391 pKa = 4.09SAFCSGDD398 pKa = 3.46QFDD401 pKa = 5.38LEE403 pKa = 4.38LAKK406 pKa = 10.61GDD408 pKa = 4.12CEE410 pKa = 4.14PTGMVIKK417 pKa = 10.24PEE419 pKa = 3.85KK420 pKa = 10.31CEE422 pKa = 3.87RR423 pKa = 11.84TKK425 pKa = 11.38DD426 pKa = 3.35PGEE429 pKa = 4.12FKK431 pKa = 11.16LLGTTYY437 pKa = 10.59RR438 pKa = 11.84GGHH441 pKa = 5.07VFRR444 pKa = 11.84DD445 pKa = 3.58TEE447 pKa = 4.14EE448 pKa = 3.94WFKK451 pKa = 11.12LALYY455 pKa = 9.52PEE457 pKa = 4.73SSVLTLDD464 pKa = 3.19ISFTRR469 pKa = 11.84LIGLWLGGAMWDD481 pKa = 4.3KK482 pKa = 10.88RR483 pKa = 11.84FCEE486 pKa = 4.09YY487 pKa = 10.34MDD489 pKa = 4.67FFQSSYY495 pKa = 10.42PCPEE499 pKa = 4.29EE500 pKa = 4.24GWFSKK505 pKa = 10.13DD506 pKa = 3.02QKK508 pKa = 10.51RR509 pKa = 11.84WLEE512 pKa = 3.89IIYY515 pKa = 10.23SGKK518 pKa = 10.27APRR521 pKa = 11.84GWTSKK526 pKa = 10.79KK527 pKa = 9.83SLFWRR532 pKa = 11.84SIFYY536 pKa = 10.51AYY538 pKa = 10.44GG539 pKa = 3.3
MM1 pKa = 7.93EE2 pKa = 6.77DD3 pKa = 3.42FTQDD7 pKa = 3.31PTTHH11 pKa = 6.67NIVAEE16 pKa = 4.58GSHH19 pKa = 7.06LIDD22 pKa = 4.56ALHH25 pKa = 6.35LRR27 pKa = 11.84PPKK30 pKa = 10.28SRR32 pKa = 11.84SATSEE37 pKa = 4.03DD38 pKa = 3.8VVSSNFRR45 pKa = 11.84SPNLAEE51 pKa = 3.65IAKK54 pKa = 9.21YY55 pKa = 10.3GGYY58 pKa = 7.94STYY61 pKa = 11.11SSNSNTDD68 pKa = 2.97PYY70 pKa = 11.29VRR72 pKa = 11.84EE73 pKa = 3.98TLKK76 pKa = 10.98LFSRR80 pKa = 11.84DD81 pKa = 3.06TYY83 pKa = 10.88EE84 pKa = 5.05DD85 pKa = 2.74IRR87 pKa = 11.84GFTRR91 pKa = 11.84RR92 pKa = 11.84PQGTPGMYY100 pKa = 8.42TALKK104 pKa = 9.81KK105 pKa = 10.83FSGEE109 pKa = 3.94RR110 pKa = 11.84NTFGDD115 pKa = 4.73LSPSQQSSMRR125 pKa = 11.84RR126 pKa = 11.84AIGKK130 pKa = 9.39AKK132 pKa = 10.31KK133 pKa = 10.09AFKK136 pKa = 10.39LPYY139 pKa = 9.55KK140 pKa = 10.34RR141 pKa = 11.84EE142 pKa = 3.91PLDD145 pKa = 3.15WHH147 pKa = 6.48EE148 pKa = 4.21VGQFLRR154 pKa = 11.84RR155 pKa = 11.84DD156 pKa = 3.56TAAGATFMGQKK167 pKa = 10.11KK168 pKa = 9.68GDD170 pKa = 3.59VMEE173 pKa = 5.62AIYY176 pKa = 10.8HH177 pKa = 5.12EE178 pKa = 4.58ARR180 pKa = 11.84WLGHH184 pKa = 6.1RR185 pKa = 11.84MKK187 pKa = 10.64QDD189 pKa = 2.56GRR191 pKa = 11.84AGFDD195 pKa = 3.4PTRR198 pKa = 11.84MRR200 pKa = 11.84FPPCLAGQRR209 pKa = 11.84GGMSEE214 pKa = 3.87IDD216 pKa = 3.64DD217 pKa = 4.28PKK219 pKa = 11.01TRR221 pKa = 11.84LVWIYY226 pKa = 8.84PAEE229 pKa = 4.02MLVVEE234 pKa = 5.03GFYY237 pKa = 11.12APLMYY242 pKa = 9.66RR243 pKa = 11.84DD244 pKa = 4.75FMSDD248 pKa = 3.17PNSPMLNGKK257 pKa = 8.31SAQRR261 pKa = 11.84LYY263 pKa = 10.74TEE265 pKa = 4.78WCCKK269 pKa = 10.29LRR271 pKa = 11.84DD272 pKa = 3.54GEE274 pKa = 4.38TLYY277 pKa = 11.52GIDD280 pKa = 4.38FSSFDD285 pKa = 3.59TKK287 pKa = 10.85VPAWLIRR294 pKa = 11.84VAFDD298 pKa = 4.45ILRR301 pKa = 11.84QNINFEE307 pKa = 4.4TFGGKK312 pKa = 9.0PVDD315 pKa = 3.87KK316 pKa = 10.81QDD318 pKa = 3.18AQKK321 pKa = 9.9WRR323 pKa = 11.84NVWDD327 pKa = 3.45AMVWYY332 pKa = 8.66FINTPILMPDD342 pKa = 2.44GRR344 pKa = 11.84MFRR347 pKa = 11.84KK348 pKa = 9.65FRR350 pKa = 11.84GVPSGSWWTQMIDD363 pKa = 3.35SVVNHH368 pKa = 6.47ILIDD372 pKa = 3.82YY373 pKa = 9.97LADD376 pKa = 3.6CQRR379 pKa = 11.84VEE381 pKa = 3.79IRR383 pKa = 11.84NLRR386 pKa = 11.84VLGDD390 pKa = 3.83DD391 pKa = 4.09SAFCSGDD398 pKa = 3.46QFDD401 pKa = 5.38LEE403 pKa = 4.38LAKK406 pKa = 10.61GDD408 pKa = 4.12CEE410 pKa = 4.14PTGMVIKK417 pKa = 10.24PEE419 pKa = 3.85KK420 pKa = 10.31CEE422 pKa = 3.87RR423 pKa = 11.84TKK425 pKa = 11.38DD426 pKa = 3.35PGEE429 pKa = 4.12FKK431 pKa = 11.16LLGTTYY437 pKa = 10.59RR438 pKa = 11.84GGHH441 pKa = 5.07VFRR444 pKa = 11.84DD445 pKa = 3.58TEE447 pKa = 4.14EE448 pKa = 3.94WFKK451 pKa = 11.12LALYY455 pKa = 9.52PEE457 pKa = 4.73SSVLTLDD464 pKa = 3.19ISFTRR469 pKa = 11.84LIGLWLGGAMWDD481 pKa = 4.3KK482 pKa = 10.88RR483 pKa = 11.84FCEE486 pKa = 4.09YY487 pKa = 10.34MDD489 pKa = 4.67FFQSSYY495 pKa = 10.42PCPEE499 pKa = 4.29EE500 pKa = 4.24GWFSKK505 pKa = 10.13DD506 pKa = 3.02QKK508 pKa = 10.51RR509 pKa = 11.84WLEE512 pKa = 3.89IIYY515 pKa = 10.23SGKK518 pKa = 10.27APRR521 pKa = 11.84GWTSKK526 pKa = 10.79KK527 pKa = 9.83SLFWRR532 pKa = 11.84SIFYY536 pKa = 10.51AYY538 pKa = 10.44GG539 pKa = 3.3
Molecular weight: 62.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
975 |
436 |
539 |
487.5 |
54.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.308 ± 2.015 | 1.128 ± 0.398 |
6.667 ± 0.418 | 5.333 ± 0.171 |
6.154 ± 0.023 | 7.282 ± 0.511 |
1.538 ± 0.04 | 3.385 ± 0.785 |
5.026 ± 0.806 | 7.487 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.564 ± 0.706 | 2.256 ± 0.251 |
5.744 ± 0.131 | 3.59 ± 0.32 |
7.487 ± 0.088 | 8.615 ± 1.15 |
6.051 ± 0.357 | 6.154 ± 1.523 |
2.462 ± 0.509 | 2.769 ± 0.965 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
