
Nannochloropsis gaditana
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; Eustigmatophyceae; Eustigmatales; Monodopsidaceae; Nannochloropsis
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 10819 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
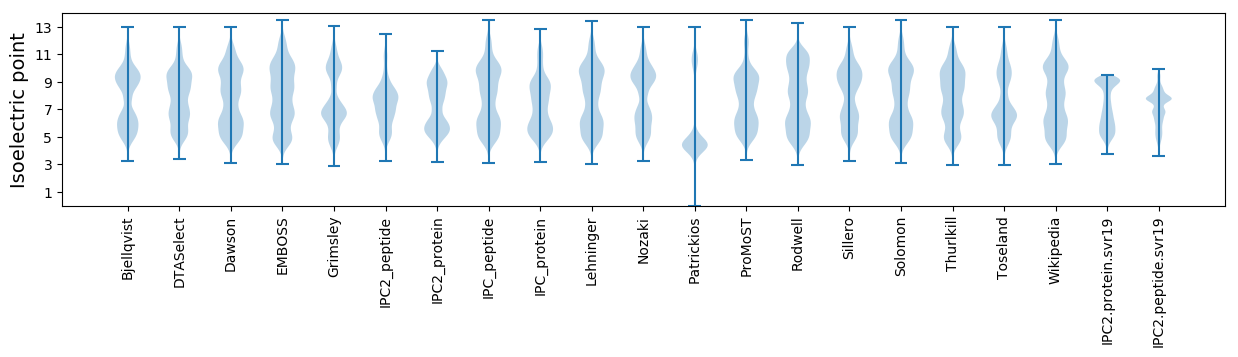
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W7T8I0|W7T8I0_9STRA Light-harvesting protein OS=Nannochloropsis gaditana OX=72520 GN=Naga_100641g3 PE=3 SV=1
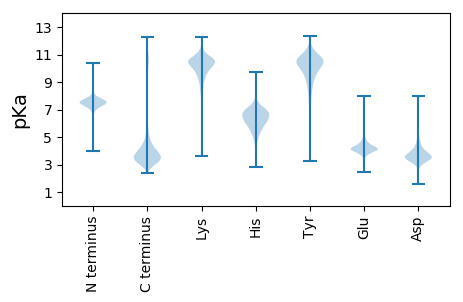
MM1 pKa = 7.54EE2 pKa = 4.54VRR4 pKa = 11.84NVLDD8 pKa = 4.55DD9 pKa = 3.4SHH11 pKa = 6.81EE12 pKa = 4.39AFDD15 pKa = 5.09CYY17 pKa = 11.34SFICIRR23 pKa = 11.84ICVFHH28 pKa = 6.74EE29 pKa = 3.94AVASNFTGAGMSVTNSVKK47 pKa = 10.57VGTGFEE53 pKa = 4.26LQSGPFMATTDD64 pKa = 3.68NFDD67 pKa = 3.5VQSDD71 pKa = 4.94DD72 pKa = 2.9ITMSYY77 pKa = 11.15SNSFQLSVTSGTAMRR92 pKa = 11.84AGDD95 pKa = 3.51ISLSEE100 pKa = 4.13AAAQIYY106 pKa = 7.56TEE108 pKa = 4.36PGVLRR113 pKa = 11.84LSTGAATFAMAARR126 pKa = 11.84GPLSVLQTKK135 pKa = 10.62ADD137 pKa = 3.64VYY139 pKa = 10.86EE140 pKa = 4.11QVADD144 pKa = 3.72EE145 pKa = 4.29WSLNSGGDD153 pKa = 3.01IDD155 pKa = 4.12VTSNGTLSMQSVSGDD170 pKa = 3.44LTFDD174 pKa = 3.54EE175 pKa = 5.25LQVEE179 pKa = 4.44TDD181 pKa = 3.18TFAIEE186 pKa = 4.29SVDD189 pKa = 3.3ILVDD193 pKa = 3.63SSEE196 pKa = 4.17TLEE199 pKa = 4.1MVVGNATTLTSPFVSHH215 pKa = 7.15FSDD218 pKa = 3.44VTLVEE223 pKa = 4.37SSEE226 pKa = 4.48SITWRR231 pKa = 11.84TGNASLSADD240 pKa = 3.21AVTIDD245 pKa = 3.49SSEE248 pKa = 4.09TLEE251 pKa = 4.1MVVGNATTLTSPFVSHH267 pKa = 7.16FSDD270 pKa = 3.37VTLPLCRR277 pKa = 11.84CGCDD281 pKa = 2.88
MM1 pKa = 7.54EE2 pKa = 4.54VRR4 pKa = 11.84NVLDD8 pKa = 4.55DD9 pKa = 3.4SHH11 pKa = 6.81EE12 pKa = 4.39AFDD15 pKa = 5.09CYY17 pKa = 11.34SFICIRR23 pKa = 11.84ICVFHH28 pKa = 6.74EE29 pKa = 3.94AVASNFTGAGMSVTNSVKK47 pKa = 10.57VGTGFEE53 pKa = 4.26LQSGPFMATTDD64 pKa = 3.68NFDD67 pKa = 3.5VQSDD71 pKa = 4.94DD72 pKa = 2.9ITMSYY77 pKa = 11.15SNSFQLSVTSGTAMRR92 pKa = 11.84AGDD95 pKa = 3.51ISLSEE100 pKa = 4.13AAAQIYY106 pKa = 7.56TEE108 pKa = 4.36PGVLRR113 pKa = 11.84LSTGAATFAMAARR126 pKa = 11.84GPLSVLQTKK135 pKa = 10.62ADD137 pKa = 3.64VYY139 pKa = 10.86EE140 pKa = 4.11QVADD144 pKa = 3.72EE145 pKa = 4.29WSLNSGGDD153 pKa = 3.01IDD155 pKa = 4.12VTSNGTLSMQSVSGDD170 pKa = 3.44LTFDD174 pKa = 3.54EE175 pKa = 5.25LQVEE179 pKa = 4.44TDD181 pKa = 3.18TFAIEE186 pKa = 4.29SVDD189 pKa = 3.3ILVDD193 pKa = 3.63SSEE196 pKa = 4.17TLEE199 pKa = 4.1MVVGNATTLTSPFVSHH215 pKa = 7.15FSDD218 pKa = 3.44VTLVEE223 pKa = 4.37SSEE226 pKa = 4.48SITWRR231 pKa = 11.84TGNASLSADD240 pKa = 3.21AVTIDD245 pKa = 3.49SSEE248 pKa = 4.09TLEE251 pKa = 4.1MVVGNATTLTSPFVSHH267 pKa = 7.16FSDD270 pKa = 3.37VTLPLCRR277 pKa = 11.84CGCDD281 pKa = 2.88
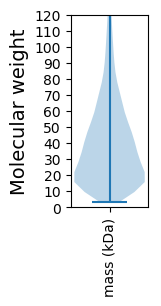
Molecular weight: 29.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W7TI78|W7TI78_9STRA Ankyrin domain protein OS=Nannochloropsis gaditana OX=72520 GN=Naga_100001g29 PE=4 SV=1
MM1 pKa = 7.13TLKK4 pKa = 9.71LTRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84WPLMTMSRR17 pKa = 11.84TMTLKK22 pKa = 9.52LTRR25 pKa = 11.84RR26 pKa = 11.84LWPLMTMSRR35 pKa = 11.84TMTLKK40 pKa = 9.23LTRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84WPPMTMSRR53 pKa = 11.84TMTSKK58 pKa = 9.24LTRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84WPLMTMSRR71 pKa = 11.84TMTSKK76 pKa = 9.24LTRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84WPLMTMSRR89 pKa = 11.84TMTLKK94 pKa = 9.23LTRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84WPPMTMSRR107 pKa = 11.84MRR109 pKa = 11.84SRR111 pKa = 11.84MMTRR115 pKa = 11.84RR116 pKa = 11.84AGMKK120 pKa = 8.58CTTKK124 pKa = 10.25RR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84LWPPMTMSRR136 pKa = 11.84MRR138 pKa = 11.84SRR140 pKa = 11.84MMTKK144 pKa = 10.07RR145 pKa = 11.84AGMKK149 pKa = 8.74CTTKK153 pKa = 10.25RR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84LWPPMTMSRR165 pKa = 11.84TRR167 pKa = 11.84SRR169 pKa = 11.84MMTRR173 pKa = 11.84RR174 pKa = 11.84AGMKK178 pKa = 8.58CTTKK182 pKa = 10.25RR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84LWPPMTMSRR194 pKa = 11.84TRR196 pKa = 11.84SRR198 pKa = 11.84MMTRR202 pKa = 11.84RR203 pKa = 11.84AGMKK207 pKa = 8.58CTTKK211 pKa = 10.25RR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84RR215 pKa = 3.54
MM1 pKa = 7.13TLKK4 pKa = 9.71LTRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84WPLMTMSRR17 pKa = 11.84TMTLKK22 pKa = 9.52LTRR25 pKa = 11.84RR26 pKa = 11.84LWPLMTMSRR35 pKa = 11.84TMTLKK40 pKa = 9.23LTRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84WPPMTMSRR53 pKa = 11.84TMTSKK58 pKa = 9.24LTRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84WPLMTMSRR71 pKa = 11.84TMTSKK76 pKa = 9.24LTRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84WPLMTMSRR89 pKa = 11.84TMTLKK94 pKa = 9.23LTRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84WPPMTMSRR107 pKa = 11.84MRR109 pKa = 11.84SRR111 pKa = 11.84MMTRR115 pKa = 11.84RR116 pKa = 11.84AGMKK120 pKa = 8.58CTTKK124 pKa = 10.25RR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84LWPPMTMSRR136 pKa = 11.84MRR138 pKa = 11.84SRR140 pKa = 11.84MMTKK144 pKa = 10.07RR145 pKa = 11.84AGMKK149 pKa = 8.74CTTKK153 pKa = 10.25RR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84LWPPMTMSRR165 pKa = 11.84TRR167 pKa = 11.84SRR169 pKa = 11.84MMTRR173 pKa = 11.84RR174 pKa = 11.84AGMKK178 pKa = 8.58CTTKK182 pKa = 10.25RR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84LWPPMTMSRR194 pKa = 11.84TRR196 pKa = 11.84SRR198 pKa = 11.84MMTRR202 pKa = 11.84RR203 pKa = 11.84AGMKK207 pKa = 8.58CTTKK211 pKa = 10.25RR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84RR215 pKa = 3.54
Molecular weight: 26.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4433024 |
29 |
5587 |
409.7 |
44.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.286 ± 0.025 | 1.617 ± 0.01 |
4.677 ± 0.016 | 6.865 ± 0.03 |
3.497 ± 0.014 | 8.533 ± 0.029 |
2.471 ± 0.011 | 3.542 ± 0.016 |
4.47 ± 0.019 | 9.844 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.288 ± 0.009 | 2.641 ± 0.014 |
6.007 ± 0.023 | 3.706 ± 0.02 |
7.248 ± 0.022 | 7.967 ± 0.029 |
5.144 ± 0.014 | 6.632 ± 0.018 |
1.308 ± 0.009 | 2.256 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
