
Sphingobium sp. Leaf26
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium; unclassified Sphingobium
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
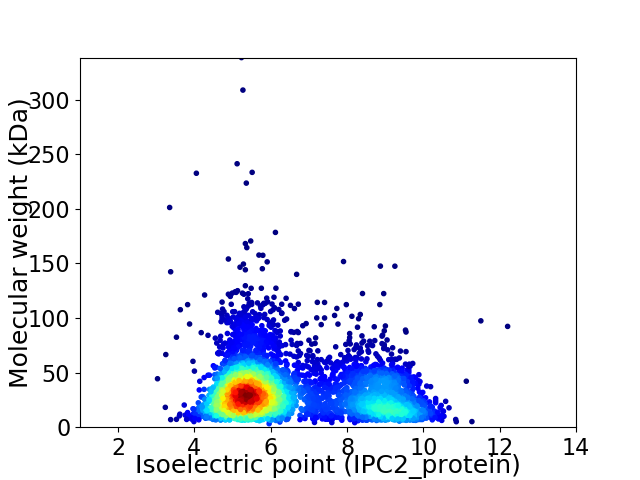
Virtual 2D-PAGE plot for 4284 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
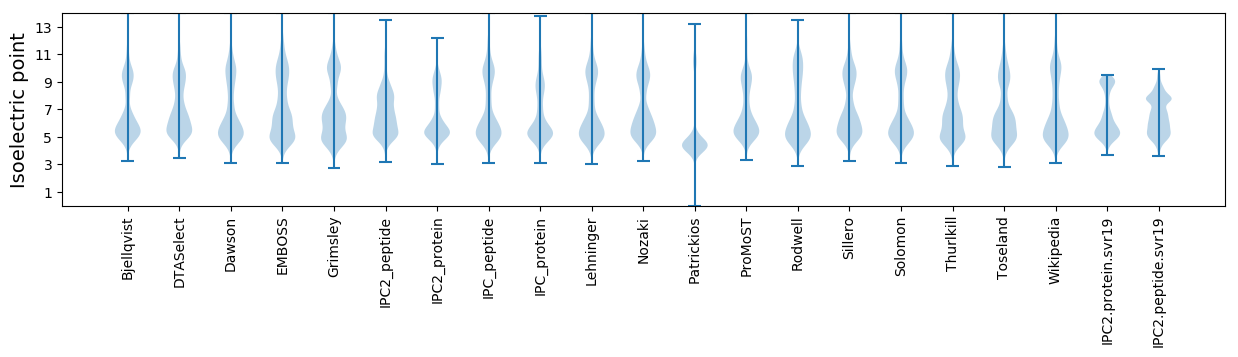
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q4J302|A0A0Q4J302_9SPHN Ubiquinone biosynthesis O-methyltransferase OS=Sphingobium sp. Leaf26 OX=1735693 GN=ubiG PE=3 SV=1
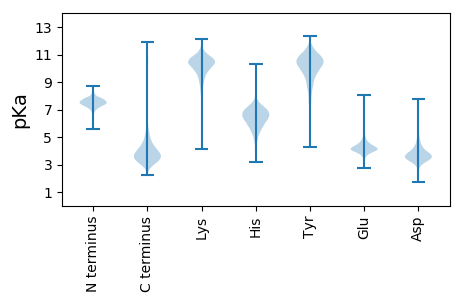
MM1 pKa = 7.69ANFSGTSGNDD11 pKa = 2.43IFTGGDD17 pKa = 3.41EE18 pKa = 5.11DD19 pKa = 5.39DD20 pKa = 4.04VATGGLGNDD29 pKa = 3.48RR30 pKa = 11.84LTGGEE35 pKa = 4.17GSDD38 pKa = 3.58ILSGDD43 pKa = 3.59EE44 pKa = 5.33GADD47 pKa = 3.46TLNGGGGDD55 pKa = 4.34DD56 pKa = 3.96YY57 pKa = 11.5LFSAGRR63 pKa = 11.84YY64 pKa = 8.43YY65 pKa = 11.05DD66 pKa = 3.88FSFSGIYY73 pKa = 9.8YY74 pKa = 10.0LNKK77 pKa = 9.68PVLDD81 pKa = 4.04TGAEE85 pKa = 4.02ADD87 pKa = 4.01TVLGGNGDD95 pKa = 3.98DD96 pKa = 5.26VIFAGYY102 pKa = 10.57NDD104 pKa = 4.69VIDD107 pKa = 4.49GGAGGEE113 pKa = 4.07DD114 pKa = 3.03TLYY117 pKa = 10.68FSLMGATHH125 pKa = 6.96GVTMDD130 pKa = 3.5FRR132 pKa = 11.84LPVIANGTGSISGIEE147 pKa = 4.04NIGWVQGSNYY157 pKa = 10.2GDD159 pKa = 3.67TLILDD164 pKa = 3.85QQGSYY169 pKa = 11.39GPFGVVYY176 pKa = 10.95AMGGDD181 pKa = 3.72DD182 pKa = 4.65HH183 pKa = 8.17VVASYY188 pKa = 8.97YY189 pKa = 9.91TDD191 pKa = 4.12HH192 pKa = 7.33IYY194 pKa = 11.08GGGGNDD200 pKa = 3.44ILDD203 pKa = 4.24GRR205 pKa = 11.84PSGYY209 pKa = 10.65LDD211 pKa = 3.69ILDD214 pKa = 4.85GGDD217 pKa = 4.15GNDD220 pKa = 3.38TLYY223 pKa = 11.47SNDD226 pKa = 3.5GSGEE230 pKa = 4.16AYY232 pKa = 10.19GGNGDD237 pKa = 3.64DD238 pKa = 4.02TIYY241 pKa = 11.06AGGLVRR247 pKa = 11.84GGAGNDD253 pKa = 3.64RR254 pKa = 11.84IVMQSSYY261 pKa = 9.34YY262 pKa = 9.71HH263 pKa = 6.24SGVFGDD269 pKa = 4.16EE270 pKa = 4.26GNDD273 pKa = 3.53EE274 pKa = 4.06IRR276 pKa = 11.84TSRR279 pKa = 11.84EE280 pKa = 3.72GNVVSGGSGADD291 pKa = 3.54RR292 pKa = 11.84LIGDD296 pKa = 4.78AGSDD300 pKa = 3.36TLIAGDD306 pKa = 4.29YY307 pKa = 10.89LPGPYY312 pKa = 9.78YY313 pKa = 10.77VGSHH317 pKa = 6.97DD318 pKa = 4.04MGTEE322 pKa = 3.83RR323 pKa = 11.84DD324 pKa = 3.93VLSGDD329 pKa = 3.66GGDD332 pKa = 4.6DD333 pKa = 3.51ILWAGWGDD341 pKa = 4.04SVDD344 pKa = 3.96GGSGTDD350 pKa = 2.82RR351 pKa = 11.84LYY353 pKa = 11.33YY354 pKa = 10.57SLGGATAGVALSTGIFASASPQALGGGTVQNVEE387 pKa = 4.2RR388 pKa = 11.84LVEE391 pKa = 4.03LMGSAFADD399 pKa = 4.92RR400 pKa = 11.84ITVATQDD407 pKa = 3.39SLLTIHH413 pKa = 7.18AGAGDD418 pKa = 4.07DD419 pKa = 4.07VVTTGDD425 pKa = 3.96SSVTLNGGDD434 pKa = 3.64GNDD437 pKa = 3.39RR438 pKa = 11.84LVSGAAADD446 pKa = 4.09SFDD449 pKa = 3.71GGAGVDD455 pKa = 3.85TIDD458 pKa = 3.32YY459 pKa = 9.44DD460 pKa = 4.07AYY462 pKa = 10.87ASGVSVNLLVGKK474 pKa = 10.01GAGGDD479 pKa = 3.41SLRR482 pKa = 11.84NVEE485 pKa = 4.81NIVGSAFADD494 pKa = 3.66TLVGDD499 pKa = 4.59GSANLITAGAGNDD512 pKa = 3.96SLDD515 pKa = 3.75GGAGADD521 pKa = 3.57RR522 pKa = 11.84LVGGLGNDD530 pKa = 3.25TYY532 pKa = 11.79YY533 pKa = 10.99VDD535 pKa = 3.66NVGDD539 pKa = 3.89VVVEE543 pKa = 4.09QAGEE547 pKa = 4.37GYY549 pKa = 10.73DD550 pKa = 3.88VVITNIDD557 pKa = 3.46YY558 pKa = 9.28TLGANVEE565 pKa = 4.53GIKK568 pKa = 10.57LLNLTKK574 pKa = 10.46GVEE577 pKa = 3.93VHH579 pKa = 7.0GNGLNNSVNAAYY591 pKa = 10.29ASAPLAAGTQIRR603 pKa = 11.84MFGEE607 pKa = 4.76GGDD610 pKa = 3.84DD611 pKa = 3.31TLLGSRR617 pKa = 11.84HH618 pKa = 6.96DD619 pKa = 4.74DD620 pKa = 3.63YY621 pKa = 11.91LDD623 pKa = 3.81GGTGVDD629 pKa = 3.68TMTGGLGNDD638 pKa = 3.57SYY640 pKa = 12.02VVDD643 pKa = 4.22DD644 pKa = 4.38ARR646 pKa = 11.84DD647 pKa = 3.7VVVEE651 pKa = 3.99QAGQGYY657 pKa = 9.9DD658 pKa = 3.5VVITSIDD665 pKa = 3.53YY666 pKa = 8.84TLGANVEE673 pKa = 4.54GIKK676 pKa = 10.47LLNVTRR682 pKa = 11.84GVEE685 pKa = 3.8VHH687 pKa = 6.67GNSLNNSFNAAYY699 pKa = 10.33VGASLAAGTQIRR711 pKa = 11.84MFGEE715 pKa = 4.63GGNDD719 pKa = 3.09TLLGSRR725 pKa = 11.84HH726 pKa = 6.08GDD728 pKa = 3.62YY729 pKa = 11.15LDD731 pKa = 4.14GGAGIDD737 pKa = 3.56TMTGGFGNDD746 pKa = 3.08TYY748 pKa = 11.83VVDD751 pKa = 3.81NKK753 pKa = 11.02GDD755 pKa = 3.76VVVEE759 pKa = 4.04QAGQGYY765 pKa = 10.09DD766 pKa = 3.49VVITNIDD773 pKa = 3.46YY774 pKa = 9.28TLGANVEE781 pKa = 4.53GIKK784 pKa = 10.54LLNLTRR790 pKa = 11.84GVEE793 pKa = 3.91VHH795 pKa = 6.92GNGLNNSLNAAYY807 pKa = 10.48ANGPLAAGTKK817 pKa = 9.24IQMFGEE823 pKa = 4.77GGNDD827 pKa = 3.14TLLGSRR833 pKa = 11.84NDD835 pKa = 4.08DD836 pKa = 3.75YY837 pKa = 11.92LDD839 pKa = 3.95GGTGVDD845 pKa = 3.42TMTGGFGNDD854 pKa = 3.02TYY856 pKa = 11.78VVDD859 pKa = 3.77NAGDD863 pKa = 3.5IVVEE867 pKa = 4.15QGGQGYY873 pKa = 9.88DD874 pKa = 3.33VVVTSIDD881 pKa = 3.6YY882 pKa = 9.39MLGANVEE889 pKa = 4.22GLKK892 pKa = 10.68LLNLTKK898 pKa = 10.55GVEE901 pKa = 3.87AHH903 pKa = 7.05GNGLNNSLNAAYY915 pKa = 10.91VNTPLAAGTTIRR927 pKa = 11.84LFGEE931 pKa = 4.68GGSDD935 pKa = 3.21SLLGSRR941 pKa = 11.84YY942 pKa = 10.36GDD944 pKa = 3.75FLDD947 pKa = 5.22GGAGNDD953 pKa = 3.67TLQGGGGNDD962 pKa = 2.98RR963 pKa = 11.84FHH965 pKa = 7.65FGDD968 pKa = 4.07GLHH971 pKa = 6.73ALSNVDD977 pKa = 4.04TILDD981 pKa = 3.79FQLGDD986 pKa = 3.86MIEE989 pKa = 4.7LDD991 pKa = 3.4DD992 pKa = 4.88AVFNGLSTGALGAGAFVANAAGAATSAEE1020 pKa = 3.73QRR1022 pKa = 11.84IVYY1025 pKa = 8.28DD1026 pKa = 3.62TDD1028 pKa = 3.28SGQLFFDD1035 pKa = 4.6ADD1037 pKa = 3.55GSGAGGAVLFAVLNGHH1053 pKa = 6.23PAIGAADD1060 pKa = 3.72FVVVV1064 pKa = 4.09
MM1 pKa = 7.69ANFSGTSGNDD11 pKa = 2.43IFTGGDD17 pKa = 3.41EE18 pKa = 5.11DD19 pKa = 5.39DD20 pKa = 4.04VATGGLGNDD29 pKa = 3.48RR30 pKa = 11.84LTGGEE35 pKa = 4.17GSDD38 pKa = 3.58ILSGDD43 pKa = 3.59EE44 pKa = 5.33GADD47 pKa = 3.46TLNGGGGDD55 pKa = 4.34DD56 pKa = 3.96YY57 pKa = 11.5LFSAGRR63 pKa = 11.84YY64 pKa = 8.43YY65 pKa = 11.05DD66 pKa = 3.88FSFSGIYY73 pKa = 9.8YY74 pKa = 10.0LNKK77 pKa = 9.68PVLDD81 pKa = 4.04TGAEE85 pKa = 4.02ADD87 pKa = 4.01TVLGGNGDD95 pKa = 3.98DD96 pKa = 5.26VIFAGYY102 pKa = 10.57NDD104 pKa = 4.69VIDD107 pKa = 4.49GGAGGEE113 pKa = 4.07DD114 pKa = 3.03TLYY117 pKa = 10.68FSLMGATHH125 pKa = 6.96GVTMDD130 pKa = 3.5FRR132 pKa = 11.84LPVIANGTGSISGIEE147 pKa = 4.04NIGWVQGSNYY157 pKa = 10.2GDD159 pKa = 3.67TLILDD164 pKa = 3.85QQGSYY169 pKa = 11.39GPFGVVYY176 pKa = 10.95AMGGDD181 pKa = 3.72DD182 pKa = 4.65HH183 pKa = 8.17VVASYY188 pKa = 8.97YY189 pKa = 9.91TDD191 pKa = 4.12HH192 pKa = 7.33IYY194 pKa = 11.08GGGGNDD200 pKa = 3.44ILDD203 pKa = 4.24GRR205 pKa = 11.84PSGYY209 pKa = 10.65LDD211 pKa = 3.69ILDD214 pKa = 4.85GGDD217 pKa = 4.15GNDD220 pKa = 3.38TLYY223 pKa = 11.47SNDD226 pKa = 3.5GSGEE230 pKa = 4.16AYY232 pKa = 10.19GGNGDD237 pKa = 3.64DD238 pKa = 4.02TIYY241 pKa = 11.06AGGLVRR247 pKa = 11.84GGAGNDD253 pKa = 3.64RR254 pKa = 11.84IVMQSSYY261 pKa = 9.34YY262 pKa = 9.71HH263 pKa = 6.24SGVFGDD269 pKa = 4.16EE270 pKa = 4.26GNDD273 pKa = 3.53EE274 pKa = 4.06IRR276 pKa = 11.84TSRR279 pKa = 11.84EE280 pKa = 3.72GNVVSGGSGADD291 pKa = 3.54RR292 pKa = 11.84LIGDD296 pKa = 4.78AGSDD300 pKa = 3.36TLIAGDD306 pKa = 4.29YY307 pKa = 10.89LPGPYY312 pKa = 9.78YY313 pKa = 10.77VGSHH317 pKa = 6.97DD318 pKa = 4.04MGTEE322 pKa = 3.83RR323 pKa = 11.84DD324 pKa = 3.93VLSGDD329 pKa = 3.66GGDD332 pKa = 4.6DD333 pKa = 3.51ILWAGWGDD341 pKa = 4.04SVDD344 pKa = 3.96GGSGTDD350 pKa = 2.82RR351 pKa = 11.84LYY353 pKa = 11.33YY354 pKa = 10.57SLGGATAGVALSTGIFASASPQALGGGTVQNVEE387 pKa = 4.2RR388 pKa = 11.84LVEE391 pKa = 4.03LMGSAFADD399 pKa = 4.92RR400 pKa = 11.84ITVATQDD407 pKa = 3.39SLLTIHH413 pKa = 7.18AGAGDD418 pKa = 4.07DD419 pKa = 4.07VVTTGDD425 pKa = 3.96SSVTLNGGDD434 pKa = 3.64GNDD437 pKa = 3.39RR438 pKa = 11.84LVSGAAADD446 pKa = 4.09SFDD449 pKa = 3.71GGAGVDD455 pKa = 3.85TIDD458 pKa = 3.32YY459 pKa = 9.44DD460 pKa = 4.07AYY462 pKa = 10.87ASGVSVNLLVGKK474 pKa = 10.01GAGGDD479 pKa = 3.41SLRR482 pKa = 11.84NVEE485 pKa = 4.81NIVGSAFADD494 pKa = 3.66TLVGDD499 pKa = 4.59GSANLITAGAGNDD512 pKa = 3.96SLDD515 pKa = 3.75GGAGADD521 pKa = 3.57RR522 pKa = 11.84LVGGLGNDD530 pKa = 3.25TYY532 pKa = 11.79YY533 pKa = 10.99VDD535 pKa = 3.66NVGDD539 pKa = 3.89VVVEE543 pKa = 4.09QAGEE547 pKa = 4.37GYY549 pKa = 10.73DD550 pKa = 3.88VVITNIDD557 pKa = 3.46YY558 pKa = 9.28TLGANVEE565 pKa = 4.53GIKK568 pKa = 10.57LLNLTKK574 pKa = 10.46GVEE577 pKa = 3.93VHH579 pKa = 7.0GNGLNNSVNAAYY591 pKa = 10.29ASAPLAAGTQIRR603 pKa = 11.84MFGEE607 pKa = 4.76GGDD610 pKa = 3.84DD611 pKa = 3.31TLLGSRR617 pKa = 11.84HH618 pKa = 6.96DD619 pKa = 4.74DD620 pKa = 3.63YY621 pKa = 11.91LDD623 pKa = 3.81GGTGVDD629 pKa = 3.68TMTGGLGNDD638 pKa = 3.57SYY640 pKa = 12.02VVDD643 pKa = 4.22DD644 pKa = 4.38ARR646 pKa = 11.84DD647 pKa = 3.7VVVEE651 pKa = 3.99QAGQGYY657 pKa = 9.9DD658 pKa = 3.5VVITSIDD665 pKa = 3.53YY666 pKa = 8.84TLGANVEE673 pKa = 4.54GIKK676 pKa = 10.47LLNVTRR682 pKa = 11.84GVEE685 pKa = 3.8VHH687 pKa = 6.67GNSLNNSFNAAYY699 pKa = 10.33VGASLAAGTQIRR711 pKa = 11.84MFGEE715 pKa = 4.63GGNDD719 pKa = 3.09TLLGSRR725 pKa = 11.84HH726 pKa = 6.08GDD728 pKa = 3.62YY729 pKa = 11.15LDD731 pKa = 4.14GGAGIDD737 pKa = 3.56TMTGGFGNDD746 pKa = 3.08TYY748 pKa = 11.83VVDD751 pKa = 3.81NKK753 pKa = 11.02GDD755 pKa = 3.76VVVEE759 pKa = 4.04QAGQGYY765 pKa = 10.09DD766 pKa = 3.49VVITNIDD773 pKa = 3.46YY774 pKa = 9.28TLGANVEE781 pKa = 4.53GIKK784 pKa = 10.54LLNLTRR790 pKa = 11.84GVEE793 pKa = 3.91VHH795 pKa = 6.92GNGLNNSLNAAYY807 pKa = 10.48ANGPLAAGTKK817 pKa = 9.24IQMFGEE823 pKa = 4.77GGNDD827 pKa = 3.14TLLGSRR833 pKa = 11.84NDD835 pKa = 4.08DD836 pKa = 3.75YY837 pKa = 11.92LDD839 pKa = 3.95GGTGVDD845 pKa = 3.42TMTGGFGNDD854 pKa = 3.02TYY856 pKa = 11.78VVDD859 pKa = 3.77NAGDD863 pKa = 3.5IVVEE867 pKa = 4.15QGGQGYY873 pKa = 9.88DD874 pKa = 3.33VVVTSIDD881 pKa = 3.6YY882 pKa = 9.39MLGANVEE889 pKa = 4.22GLKK892 pKa = 10.68LLNLTKK898 pKa = 10.55GVEE901 pKa = 3.87AHH903 pKa = 7.05GNGLNNSLNAAYY915 pKa = 10.91VNTPLAAGTTIRR927 pKa = 11.84LFGEE931 pKa = 4.68GGSDD935 pKa = 3.21SLLGSRR941 pKa = 11.84YY942 pKa = 10.36GDD944 pKa = 3.75FLDD947 pKa = 5.22GGAGNDD953 pKa = 3.67TLQGGGGNDD962 pKa = 2.98RR963 pKa = 11.84FHH965 pKa = 7.65FGDD968 pKa = 4.07GLHH971 pKa = 6.73ALSNVDD977 pKa = 4.04TILDD981 pKa = 3.79FQLGDD986 pKa = 3.86MIEE989 pKa = 4.7LDD991 pKa = 3.4DD992 pKa = 4.88AVFNGLSTGALGAGAFVANAAGAATSAEE1020 pKa = 3.73QRR1022 pKa = 11.84IVYY1025 pKa = 8.28DD1026 pKa = 3.62TDD1028 pKa = 3.28SGQLFFDD1035 pKa = 4.6ADD1037 pKa = 3.55GSGAGGAVLFAVLNGHH1053 pKa = 6.23PAIGAADD1060 pKa = 3.72FVVVV1064 pKa = 4.09
Molecular weight: 107.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q4III5|A0A0Q4III5_9SPHN Uncharacterized protein OS=Sphingobium sp. Leaf26 OX=1735693 GN=ASE85_08780 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06SLSAA44 pKa = 3.93
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1403714 |
29 |
3092 |
327.7 |
35.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.055 ± 0.059 | 0.767 ± 0.01 |
6.264 ± 0.031 | 4.868 ± 0.039 |
3.527 ± 0.023 | 8.941 ± 0.042 |
2.05 ± 0.019 | 5.204 ± 0.023 |
3.048 ± 0.035 | 9.877 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.643 ± 0.038 | 2.615 ± 0.028 |
5.307 ± 0.032 | 3.337 ± 0.024 |
7.11 ± 0.041 | 5.289 ± 0.036 |
5.298 ± 0.04 | 6.976 ± 0.029 |
1.476 ± 0.017 | 2.349 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
