
Panicum hallii var. hallii
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Panicinae; Panicum; Panicum sect. Panicum;
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
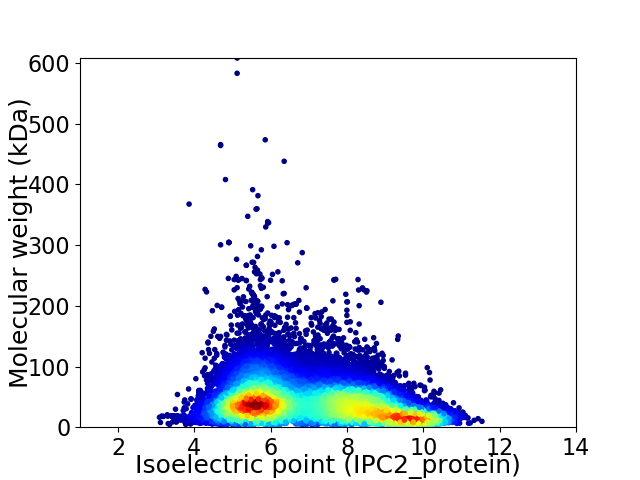
Virtual 2D-PAGE plot for 38607 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
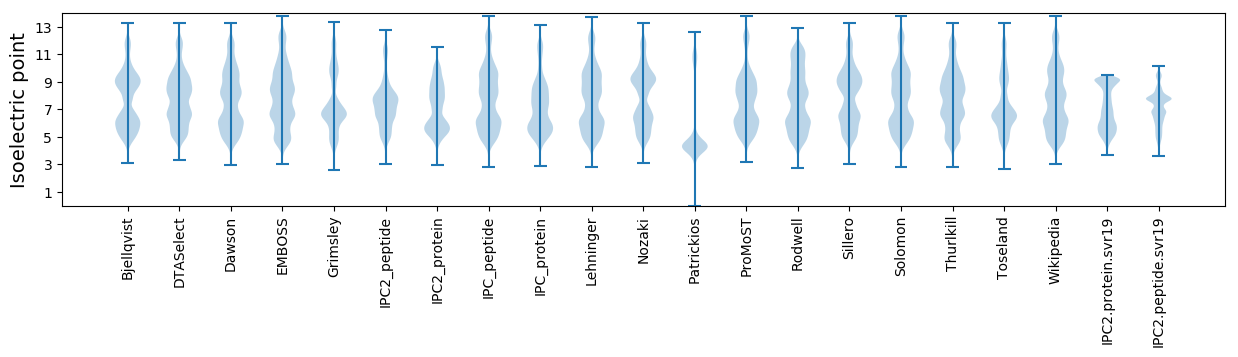
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T7EAB1|A0A2T7EAB1_9POAL MFS domain-containing protein OS=Panicum hallii var. hallii OX=1504633 GN=GQ55_3G167400 PE=3 SV=1
MM1 pKa = 8.2DD2 pKa = 4.66CTHH5 pKa = 6.45HH6 pKa = 6.28QPRR9 pKa = 11.84PTMTPNPEE17 pKa = 4.24PSPAPAPPTAAPQEE31 pKa = 4.13ATGDD35 pKa = 4.01SEE37 pKa = 4.9SDD39 pKa = 2.97SDD41 pKa = 4.08TTVSALLTGDD51 pKa = 3.82HH52 pKa = 7.06PSVPGYY58 pKa = 10.65DD59 pKa = 4.22DD60 pKa = 4.61EE61 pKa = 7.83DD62 pKa = 4.23DD63 pKa = 5.11AEE65 pKa = 4.48SCSGGDD71 pKa = 3.93GGSGLCGGATSAAGGDD87 pKa = 4.02GSDD90 pKa = 4.06GDD92 pKa = 4.55DD93 pKa = 3.63DD94 pKa = 4.27VGAEE98 pKa = 3.84RR99 pKa = 11.84GEE101 pKa = 4.43AEE103 pKa = 4.58VDD105 pKa = 3.14SSMAVPWWRR114 pKa = 11.84RR115 pKa = 11.84MAVEE119 pKa = 4.14EE120 pKa = 3.93AAAAAAHH127 pKa = 7.5DD128 pKa = 4.57DD129 pKa = 4.27GGWFAPAAEE138 pKa = 4.49AGVVAGAGGHH148 pKa = 5.1SAEE151 pKa = 4.41SNILFWEE158 pKa = 4.3ACIEE162 pKa = 3.98HH163 pKa = 7.12GYY165 pKa = 10.49
MM1 pKa = 8.2DD2 pKa = 4.66CTHH5 pKa = 6.45HH6 pKa = 6.28QPRR9 pKa = 11.84PTMTPNPEE17 pKa = 4.24PSPAPAPPTAAPQEE31 pKa = 4.13ATGDD35 pKa = 4.01SEE37 pKa = 4.9SDD39 pKa = 2.97SDD41 pKa = 4.08TTVSALLTGDD51 pKa = 3.82HH52 pKa = 7.06PSVPGYY58 pKa = 10.65DD59 pKa = 4.22DD60 pKa = 4.61EE61 pKa = 7.83DD62 pKa = 4.23DD63 pKa = 5.11AEE65 pKa = 4.48SCSGGDD71 pKa = 3.93GGSGLCGGATSAAGGDD87 pKa = 4.02GSDD90 pKa = 4.06GDD92 pKa = 4.55DD93 pKa = 3.63DD94 pKa = 4.27VGAEE98 pKa = 3.84RR99 pKa = 11.84GEE101 pKa = 4.43AEE103 pKa = 4.58VDD105 pKa = 3.14SSMAVPWWRR114 pKa = 11.84RR115 pKa = 11.84MAVEE119 pKa = 4.14EE120 pKa = 3.93AAAAAAHH127 pKa = 7.5DD128 pKa = 4.57DD129 pKa = 4.27GGWFAPAAEE138 pKa = 4.49AGVVAGAGGHH148 pKa = 5.1SAEE151 pKa = 4.41SNILFWEE158 pKa = 4.3ACIEE162 pKa = 3.98HH163 pKa = 7.12GYY165 pKa = 10.49
Molecular weight: 16.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T7EWV5|A0A2T7EWV5_9POAL Uncharacterized protein OS=Panicum hallii var. hallii OX=1504633 GN=GQ55_2G383200 PE=4 SV=1
MM1 pKa = 7.65ARR3 pKa = 11.84RR4 pKa = 11.84GWARR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84GRR12 pKa = 11.84AWPPGLARR20 pKa = 11.84RR21 pKa = 11.84HH22 pKa = 4.08GWARR26 pKa = 11.84RR27 pKa = 11.84PAWAQRR33 pKa = 11.84GSAQWVMARR42 pKa = 11.84RR43 pKa = 11.84GWARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84AWPPGLARR59 pKa = 11.84RR60 pKa = 11.84HH61 pKa = 4.08GWARR65 pKa = 11.84RR66 pKa = 11.84PAWARR71 pKa = 11.84WPGKK75 pKa = 10.0ARR77 pKa = 11.84QPGKK81 pKa = 9.43AWRR84 pKa = 11.84RR85 pKa = 11.84GWARR89 pKa = 11.84RR90 pKa = 11.84SGSLACC96 pKa = 5.39
MM1 pKa = 7.65ARR3 pKa = 11.84RR4 pKa = 11.84GWARR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84GRR12 pKa = 11.84AWPPGLARR20 pKa = 11.84RR21 pKa = 11.84HH22 pKa = 4.08GWARR26 pKa = 11.84RR27 pKa = 11.84PAWAQRR33 pKa = 11.84GSAQWVMARR42 pKa = 11.84RR43 pKa = 11.84GWARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84AWPPGLARR59 pKa = 11.84RR60 pKa = 11.84HH61 pKa = 4.08GWARR65 pKa = 11.84RR66 pKa = 11.84PAWARR71 pKa = 11.84WPGKK75 pKa = 10.0ARR77 pKa = 11.84QPGKK81 pKa = 9.43AWRR84 pKa = 11.84RR85 pKa = 11.84GWARR89 pKa = 11.84RR90 pKa = 11.84SGSLACC96 pKa = 5.39
Molecular weight: 11.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
15483065 |
29 |
5381 |
401.0 |
44.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.453 ± 0.019 | 1.923 ± 0.007 |
5.309 ± 0.008 | 6.004 ± 0.014 |
3.617 ± 0.008 | 7.358 ± 0.015 |
2.464 ± 0.006 | 4.29 ± 0.009 |
4.867 ± 0.011 | 9.491 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.005 | 3.458 ± 0.009 |
5.787 ± 0.013 | 3.543 ± 0.01 |
6.483 ± 0.013 | 8.38 ± 0.016 |
4.715 ± 0.007 | 6.652 ± 0.009 |
1.296 ± 0.005 | 2.537 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
