
Desulfotomaculum arcticum DSM 17038
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Desulfallaceae; Desulfallas; Desulfallas arcticus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
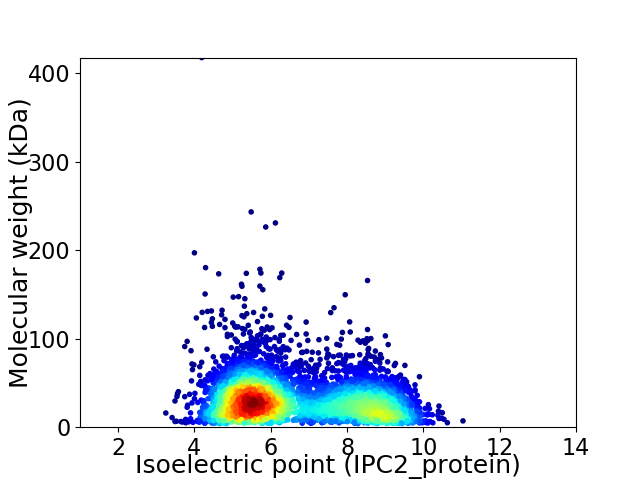
Virtual 2D-PAGE plot for 5051 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2PJJ3|A0A1I2PJJ3_9FIRM ABC-2 type transport system permease protein OS=Desulfotomaculum arcticum DSM 17038 OX=1121424 GN=SAMN05660649_00907 PE=4 SV=1
MM1 pKa = 7.23YY2 pKa = 10.32IFVKK6 pKa = 10.48GYY8 pKa = 10.71NLAKK12 pKa = 10.38DD13 pKa = 3.51RR14 pKa = 11.84NGRR17 pKa = 11.84LINIIVRR24 pKa = 11.84TLRR27 pKa = 11.84KK28 pKa = 9.61YY29 pKa = 7.68IHH31 pKa = 7.01KK32 pKa = 10.61YY33 pKa = 10.45GILFLLLCLFSTMTACEE50 pKa = 4.24SKK52 pKa = 10.81DD53 pKa = 3.15ILGDD57 pKa = 3.76DD58 pKa = 4.11EE59 pKa = 5.84PEE61 pKa = 4.0VSQNATEE68 pKa = 4.68DD69 pKa = 3.47GTMGSSNDD77 pKa = 3.69GVNTEE82 pKa = 4.06ADD84 pKa = 3.26KK85 pKa = 11.38GAEE88 pKa = 3.83QSTAPNEE95 pKa = 4.22LEE97 pKa = 4.33DD98 pKa = 4.14TVSGQDD104 pKa = 3.96GEE106 pKa = 4.38EE107 pKa = 4.12TQMNEE112 pKa = 3.52ADD114 pKa = 3.65IDD116 pKa = 3.85RR117 pKa = 11.84EE118 pKa = 4.27AMMNEE123 pKa = 4.07YY124 pKa = 10.69KK125 pKa = 10.76SILKK129 pKa = 10.36GIYY132 pKa = 9.44DD133 pKa = 3.5NHH135 pKa = 6.7IFPDD139 pKa = 4.37GQACDD144 pKa = 3.13WDD146 pKa = 3.92GRR148 pKa = 11.84SDD150 pKa = 3.44VTQNQFAVYY159 pKa = 10.22DD160 pKa = 4.02IDD162 pKa = 5.61QDD164 pKa = 4.09TQDD167 pKa = 4.34EE168 pKa = 5.12LILAYY173 pKa = 9.41TNSSMAGMIEE183 pKa = 5.15LIYY186 pKa = 11.14DD187 pKa = 3.95FDD189 pKa = 5.41NDD191 pKa = 3.38TKK193 pKa = 10.56TANEE197 pKa = 4.12EE198 pKa = 3.85FRR200 pKa = 11.84EE201 pKa = 4.46FPTLTYY207 pKa = 10.19YY208 pKa = 11.24DD209 pKa = 3.58NGIIKK214 pKa = 10.32ADD216 pKa = 3.23WSHH219 pKa = 5.59NQGLAGDD226 pKa = 4.19FWPYY230 pKa = 10.32TLYY233 pKa = 10.77QYY235 pKa = 11.03DD236 pKa = 4.35KK237 pKa = 11.11KK238 pKa = 10.93SDD240 pKa = 3.6SYY242 pKa = 11.84VDD244 pKa = 3.4VGMVDD249 pKa = 2.56AWDD252 pKa = 4.23RR253 pKa = 11.84SLSATDD259 pKa = 3.7YY260 pKa = 11.4NGDD263 pKa = 3.85PFPAEE268 pKa = 4.22SDD270 pKa = 3.3TDD272 pKa = 3.76GDD274 pKa = 4.34GIVYY278 pKa = 10.27YY279 pKa = 10.31IMQDD283 pKa = 3.06GEE285 pKa = 4.6YY286 pKa = 10.61KK287 pKa = 10.23LDD289 pKa = 3.69HH290 pKa = 7.17PIDD293 pKa = 3.96GAEE296 pKa = 3.87YY297 pKa = 8.46EE298 pKa = 4.16QWRR301 pKa = 11.84DD302 pKa = 3.46FYY304 pKa = 10.92IGNAKK309 pKa = 9.28EE310 pKa = 3.97VNIPFINLTEE320 pKa = 4.02EE321 pKa = 4.45NIDD324 pKa = 3.97NIQQ327 pKa = 3.01
MM1 pKa = 7.23YY2 pKa = 10.32IFVKK6 pKa = 10.48GYY8 pKa = 10.71NLAKK12 pKa = 10.38DD13 pKa = 3.51RR14 pKa = 11.84NGRR17 pKa = 11.84LINIIVRR24 pKa = 11.84TLRR27 pKa = 11.84KK28 pKa = 9.61YY29 pKa = 7.68IHH31 pKa = 7.01KK32 pKa = 10.61YY33 pKa = 10.45GILFLLLCLFSTMTACEE50 pKa = 4.24SKK52 pKa = 10.81DD53 pKa = 3.15ILGDD57 pKa = 3.76DD58 pKa = 4.11EE59 pKa = 5.84PEE61 pKa = 4.0VSQNATEE68 pKa = 4.68DD69 pKa = 3.47GTMGSSNDD77 pKa = 3.69GVNTEE82 pKa = 4.06ADD84 pKa = 3.26KK85 pKa = 11.38GAEE88 pKa = 3.83QSTAPNEE95 pKa = 4.22LEE97 pKa = 4.33DD98 pKa = 4.14TVSGQDD104 pKa = 3.96GEE106 pKa = 4.38EE107 pKa = 4.12TQMNEE112 pKa = 3.52ADD114 pKa = 3.65IDD116 pKa = 3.85RR117 pKa = 11.84EE118 pKa = 4.27AMMNEE123 pKa = 4.07YY124 pKa = 10.69KK125 pKa = 10.76SILKK129 pKa = 10.36GIYY132 pKa = 9.44DD133 pKa = 3.5NHH135 pKa = 6.7IFPDD139 pKa = 4.37GQACDD144 pKa = 3.13WDD146 pKa = 3.92GRR148 pKa = 11.84SDD150 pKa = 3.44VTQNQFAVYY159 pKa = 10.22DD160 pKa = 4.02IDD162 pKa = 5.61QDD164 pKa = 4.09TQDD167 pKa = 4.34EE168 pKa = 5.12LILAYY173 pKa = 9.41TNSSMAGMIEE183 pKa = 5.15LIYY186 pKa = 11.14DD187 pKa = 3.95FDD189 pKa = 5.41NDD191 pKa = 3.38TKK193 pKa = 10.56TANEE197 pKa = 4.12EE198 pKa = 3.85FRR200 pKa = 11.84EE201 pKa = 4.46FPTLTYY207 pKa = 10.19YY208 pKa = 11.24DD209 pKa = 3.58NGIIKK214 pKa = 10.32ADD216 pKa = 3.23WSHH219 pKa = 5.59NQGLAGDD226 pKa = 4.19FWPYY230 pKa = 10.32TLYY233 pKa = 10.77QYY235 pKa = 11.03DD236 pKa = 4.35KK237 pKa = 11.11KK238 pKa = 10.93SDD240 pKa = 3.6SYY242 pKa = 11.84VDD244 pKa = 3.4VGMVDD249 pKa = 2.56AWDD252 pKa = 4.23RR253 pKa = 11.84SLSATDD259 pKa = 3.7YY260 pKa = 11.4NGDD263 pKa = 3.85PFPAEE268 pKa = 4.22SDD270 pKa = 3.3TDD272 pKa = 3.76GDD274 pKa = 4.34GIVYY278 pKa = 10.27YY279 pKa = 10.31IMQDD283 pKa = 3.06GEE285 pKa = 4.6YY286 pKa = 10.61KK287 pKa = 10.23LDD289 pKa = 3.69HH290 pKa = 7.17PIDD293 pKa = 3.96GAEE296 pKa = 3.87YY297 pKa = 8.46EE298 pKa = 4.16QWRR301 pKa = 11.84DD302 pKa = 3.46FYY304 pKa = 10.92IGNAKK309 pKa = 9.28EE310 pKa = 3.97VNIPFINLTEE320 pKa = 4.02EE321 pKa = 4.45NIDD324 pKa = 3.97NIQQ327 pKa = 3.01
Molecular weight: 37.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2ZC58|A0A1I2ZC58_9FIRM Transposase DDE domain-containing protein (Fragment) OS=Desulfotomaculum arcticum DSM 17038 OX=1121424 GN=SAMN05660649_04876 PE=4 SV=1
MM1 pKa = 7.68PKK3 pKa = 9.97IKK5 pKa = 8.1THH7 pKa = 6.4RR8 pKa = 11.84GAAKK12 pKa = 10.0RR13 pKa = 11.84FKK15 pKa = 9.91KK16 pKa = 9.16TGSGKK21 pKa = 10.45FKK23 pKa = 10.02TSHH26 pKa = 6.54AFHH29 pKa = 6.38SHH31 pKa = 5.49ILGKK35 pKa = 8.81KK36 pKa = 5.69TPKK39 pKa = 9.96RR40 pKa = 11.84KK41 pKa = 9.57RR42 pKa = 11.84KK43 pKa = 9.1LRR45 pKa = 11.84KK46 pKa = 9.37ALIVHH51 pKa = 6.96PANAANLQRR60 pKa = 11.84LLPP63 pKa = 4.35
MM1 pKa = 7.68PKK3 pKa = 9.97IKK5 pKa = 8.1THH7 pKa = 6.4RR8 pKa = 11.84GAAKK12 pKa = 10.0RR13 pKa = 11.84FKK15 pKa = 9.91KK16 pKa = 9.16TGSGKK21 pKa = 10.45FKK23 pKa = 10.02TSHH26 pKa = 6.54AFHH29 pKa = 6.38SHH31 pKa = 5.49ILGKK35 pKa = 8.81KK36 pKa = 5.69TPKK39 pKa = 9.96RR40 pKa = 11.84KK41 pKa = 9.57RR42 pKa = 11.84KK43 pKa = 9.1LRR45 pKa = 11.84KK46 pKa = 9.37ALIVHH51 pKa = 6.96PANAANLQRR60 pKa = 11.84LLPP63 pKa = 4.35
Molecular weight: 7.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1486842 |
39 |
3953 |
294.4 |
32.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.343 ± 0.037 | 1.232 ± 0.018 |
5.067 ± 0.034 | 6.55 ± 0.032 |
3.915 ± 0.026 | 7.754 ± 0.03 |
1.738 ± 0.017 | 7.329 ± 0.031 |
6.026 ± 0.028 | 10.057 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.718 ± 0.018 | 4.374 ± 0.027 |
4.133 ± 0.02 | 3.511 ± 0.024 |
5.002 ± 0.032 | 5.466 ± 0.023 |
5.091 ± 0.032 | 7.347 ± 0.033 |
1.015 ± 0.014 | 3.331 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
