
Vibrio phage VEJphi
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Fibrovirus; unclassified Fibrovirus
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
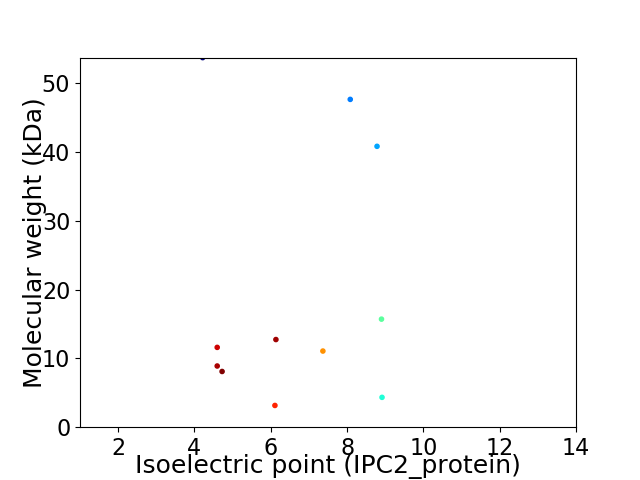
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C3W4N9|C3W4N9_9VIRU Putative minor capsid protein OS=Vibrio phage VEJphi OX=642111 PE=4 SV=1
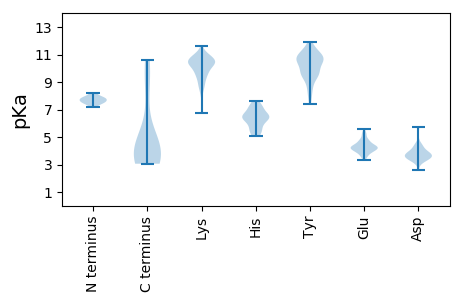
MM1 pKa = 8.22VMFMRR6 pKa = 11.84KK7 pKa = 8.88FFIISLVISLYY18 pKa = 9.72FTVLPAFAAYY28 pKa = 9.47QLPFDD33 pKa = 3.86QTKK36 pKa = 8.61TFPTVQAAAEE46 pKa = 4.38YY47 pKa = 9.69YY48 pKa = 10.96ANLLGGTSCKK58 pKa = 10.4ADD60 pKa = 3.16GNRR63 pKa = 11.84FKK65 pKa = 10.61IYY67 pKa = 9.57RR68 pKa = 11.84VKK70 pKa = 10.74SISDD74 pKa = 3.5PSFVIQQEE82 pKa = 4.58TYY84 pKa = 11.16LDD86 pKa = 3.67NKK88 pKa = 10.05CQVFSSKK95 pKa = 11.03GDD97 pKa = 3.25ISVTLIVVDD106 pKa = 5.71DD107 pKa = 4.16PTTCEE112 pKa = 3.7ASKK115 pKa = 10.83GQTGKK120 pKa = 10.93VGWNSYY126 pKa = 7.76FWGSATPSRR135 pKa = 11.84YY136 pKa = 9.22ICSSANGGCVALTGDD151 pKa = 4.48HH152 pKa = 6.64LCINIDD158 pKa = 3.65PEE160 pKa = 4.19ALKK163 pKa = 10.6NDD165 pKa = 3.8PSLYY169 pKa = 10.79DD170 pKa = 3.68CDD172 pKa = 3.91ALYY175 pKa = 10.18IVQDD179 pKa = 4.39TPCSPSGDD187 pKa = 3.77YY188 pKa = 10.5PFCTDD193 pKa = 3.63EE194 pKa = 4.45NCSSFLPEE202 pKa = 4.96PNPNPNPEE210 pKa = 4.47PEE212 pKa = 4.8PEE214 pKa = 4.76PEE216 pKa = 4.01PQPDD220 pKa = 3.99PEE222 pKa = 5.19HH223 pKa = 6.61NPSDD227 pKa = 3.78PTAPLPDD234 pKa = 3.67SGGEE238 pKa = 4.12VINPSVPPKK247 pKa = 10.39PPTEE251 pKa = 4.16EE252 pKa = 3.87KK253 pKa = 10.28PKK255 pKa = 10.44PDD257 pKa = 3.82VEE259 pKa = 4.76TPDD262 pKa = 3.77PTPDD266 pKa = 3.45SNSDD270 pKa = 3.38VVQSVTGMNEE280 pKa = 3.81DD281 pKa = 3.54MNEE284 pKa = 3.69LLTRR288 pKa = 11.84LNSDD292 pKa = 3.45NNKK295 pKa = 9.66QLDD298 pKa = 3.95DD299 pKa = 4.12VNNQLLQLNTQSQRR313 pKa = 11.84IVAQIAKK320 pKa = 9.05QEE322 pKa = 4.11KK323 pKa = 8.44QDD325 pKa = 3.42AAIYY329 pKa = 9.41EE330 pKa = 4.23NTKK333 pKa = 11.01ALIQNLNKK341 pKa = 10.62DD342 pKa = 3.06VTTAVNKK349 pKa = 6.75TTNAVNALGSKK360 pKa = 10.33VDD362 pKa = 3.84GLSDD366 pKa = 3.48AVDD369 pKa = 3.62GLGEE373 pKa = 4.32DD374 pKa = 3.59VSAIKK379 pKa = 10.5DD380 pKa = 3.59AITNVDD386 pKa = 3.12TSGAGISGTCIEE398 pKa = 5.16SDD400 pKa = 3.5TCTGFYY406 pKa = 10.59EE407 pKa = 4.24SGYY410 pKa = 9.37PDD412 pKa = 5.33GISGIFTEE420 pKa = 5.44HH421 pKa = 7.08FDD423 pKa = 3.95TVSKK427 pKa = 11.01SVTDD431 pKa = 3.68TVKK434 pKa = 11.13DD435 pKa = 3.93FMKK438 pKa = 10.12IDD440 pKa = 3.92LSHH443 pKa = 6.56AQRR446 pKa = 11.84PSFSIPVLHH455 pKa = 7.04FGNFSFDD462 pKa = 4.17DD463 pKa = 4.45YY464 pKa = 11.88INLDD468 pKa = 3.69WIFGFVRR475 pKa = 11.84VCMMVSTAFLCRR487 pKa = 11.84KK488 pKa = 9.77IIFGGG493 pKa = 3.5
MM1 pKa = 8.22VMFMRR6 pKa = 11.84KK7 pKa = 8.88FFIISLVISLYY18 pKa = 9.72FTVLPAFAAYY28 pKa = 9.47QLPFDD33 pKa = 3.86QTKK36 pKa = 8.61TFPTVQAAAEE46 pKa = 4.38YY47 pKa = 9.69YY48 pKa = 10.96ANLLGGTSCKK58 pKa = 10.4ADD60 pKa = 3.16GNRR63 pKa = 11.84FKK65 pKa = 10.61IYY67 pKa = 9.57RR68 pKa = 11.84VKK70 pKa = 10.74SISDD74 pKa = 3.5PSFVIQQEE82 pKa = 4.58TYY84 pKa = 11.16LDD86 pKa = 3.67NKK88 pKa = 10.05CQVFSSKK95 pKa = 11.03GDD97 pKa = 3.25ISVTLIVVDD106 pKa = 5.71DD107 pKa = 4.16PTTCEE112 pKa = 3.7ASKK115 pKa = 10.83GQTGKK120 pKa = 10.93VGWNSYY126 pKa = 7.76FWGSATPSRR135 pKa = 11.84YY136 pKa = 9.22ICSSANGGCVALTGDD151 pKa = 4.48HH152 pKa = 6.64LCINIDD158 pKa = 3.65PEE160 pKa = 4.19ALKK163 pKa = 10.6NDD165 pKa = 3.8PSLYY169 pKa = 10.79DD170 pKa = 3.68CDD172 pKa = 3.91ALYY175 pKa = 10.18IVQDD179 pKa = 4.39TPCSPSGDD187 pKa = 3.77YY188 pKa = 10.5PFCTDD193 pKa = 3.63EE194 pKa = 4.45NCSSFLPEE202 pKa = 4.96PNPNPNPEE210 pKa = 4.47PEE212 pKa = 4.8PEE214 pKa = 4.76PEE216 pKa = 4.01PQPDD220 pKa = 3.99PEE222 pKa = 5.19HH223 pKa = 6.61NPSDD227 pKa = 3.78PTAPLPDD234 pKa = 3.67SGGEE238 pKa = 4.12VINPSVPPKK247 pKa = 10.39PPTEE251 pKa = 4.16EE252 pKa = 3.87KK253 pKa = 10.28PKK255 pKa = 10.44PDD257 pKa = 3.82VEE259 pKa = 4.76TPDD262 pKa = 3.77PTPDD266 pKa = 3.45SNSDD270 pKa = 3.38VVQSVTGMNEE280 pKa = 3.81DD281 pKa = 3.54MNEE284 pKa = 3.69LLTRR288 pKa = 11.84LNSDD292 pKa = 3.45NNKK295 pKa = 9.66QLDD298 pKa = 3.95DD299 pKa = 4.12VNNQLLQLNTQSQRR313 pKa = 11.84IVAQIAKK320 pKa = 9.05QEE322 pKa = 4.11KK323 pKa = 8.44QDD325 pKa = 3.42AAIYY329 pKa = 9.41EE330 pKa = 4.23NTKK333 pKa = 11.01ALIQNLNKK341 pKa = 10.62DD342 pKa = 3.06VTTAVNKK349 pKa = 6.75TTNAVNALGSKK360 pKa = 10.33VDD362 pKa = 3.84GLSDD366 pKa = 3.48AVDD369 pKa = 3.62GLGEE373 pKa = 4.32DD374 pKa = 3.59VSAIKK379 pKa = 10.5DD380 pKa = 3.59AITNVDD386 pKa = 3.12TSGAGISGTCIEE398 pKa = 5.16SDD400 pKa = 3.5TCTGFYY406 pKa = 10.59EE407 pKa = 4.24SGYY410 pKa = 9.37PDD412 pKa = 5.33GISGIFTEE420 pKa = 5.44HH421 pKa = 7.08FDD423 pKa = 3.95TVSKK427 pKa = 11.01SVTDD431 pKa = 3.68TVKK434 pKa = 11.13DD435 pKa = 3.93FMKK438 pKa = 10.12IDD440 pKa = 3.92LSHH443 pKa = 6.56AQRR446 pKa = 11.84PSFSIPVLHH455 pKa = 7.04FGNFSFDD462 pKa = 4.17DD463 pKa = 4.45YY464 pKa = 11.88INLDD468 pKa = 3.69WIFGFVRR475 pKa = 11.84VCMMVSTAFLCRR487 pKa = 11.84KK488 pKa = 9.77IIFGGG493 pKa = 3.5
Molecular weight: 53.71 kDa
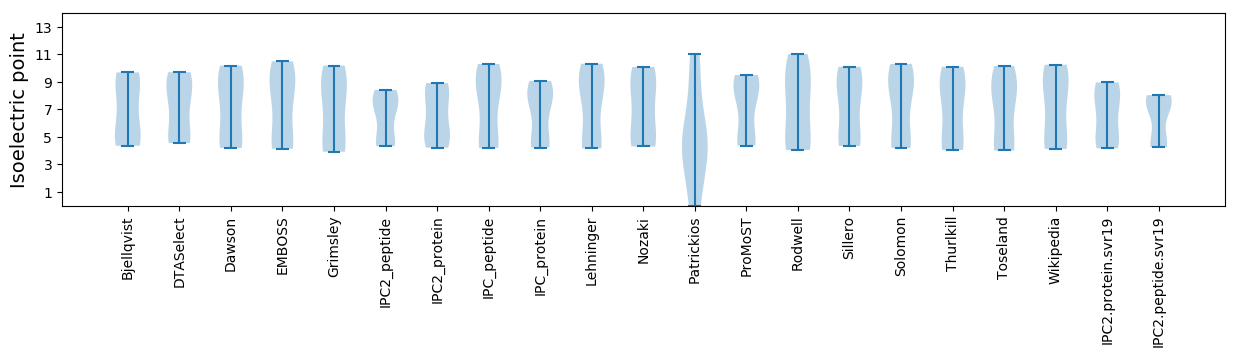
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C3W4P2|C3W4P2_9VIRU Putative regulator protein OS=Vibrio phage VEJphi OX=642111 PE=4 SV=1
MM1 pKa = 7.7LIKK4 pKa = 10.73LPILGVLSMNRR15 pKa = 11.84SEE17 pKa = 4.0MTKK20 pKa = 9.96NFVFRR25 pKa = 11.84EE26 pKa = 4.35FEE28 pKa = 4.32CGLSVEE34 pKa = 4.72EE35 pKa = 4.5AAKK38 pKa = 10.83LCFKK42 pKa = 9.77TVNEE46 pKa = 4.24VKK48 pKa = 10.31QWDD51 pKa = 4.03AGEE54 pKa = 4.75KK55 pKa = 9.98IPPICKK61 pKa = 9.68RR62 pKa = 11.84LMRR65 pKa = 11.84WHH67 pKa = 6.19SRR69 pKa = 11.84KK70 pKa = 9.28EE71 pKa = 3.83LYY73 pKa = 10.74YY74 pKa = 10.93GDD76 pKa = 4.06EE77 pKa = 3.55WWGFRR82 pKa = 11.84MEE84 pKa = 4.38GGRR87 pKa = 11.84LIFPTGDD94 pKa = 2.67RR95 pKa = 11.84VAPQQLLAAIAILQIQAPDD114 pKa = 3.52DD115 pKa = 3.8AMTRR119 pKa = 11.84SKK121 pKa = 10.76LLKK124 pKa = 9.49YY125 pKa = 9.56ARR127 pKa = 11.84AMARR131 pKa = 11.84IKK133 pKa = 10.59GIKK136 pKa = 8.89
MM1 pKa = 7.7LIKK4 pKa = 10.73LPILGVLSMNRR15 pKa = 11.84SEE17 pKa = 4.0MTKK20 pKa = 9.96NFVFRR25 pKa = 11.84EE26 pKa = 4.35FEE28 pKa = 4.32CGLSVEE34 pKa = 4.72EE35 pKa = 4.5AAKK38 pKa = 10.83LCFKK42 pKa = 9.77TVNEE46 pKa = 4.24VKK48 pKa = 10.31QWDD51 pKa = 4.03AGEE54 pKa = 4.75KK55 pKa = 9.98IPPICKK61 pKa = 9.68RR62 pKa = 11.84LMRR65 pKa = 11.84WHH67 pKa = 6.19SRR69 pKa = 11.84KK70 pKa = 9.28EE71 pKa = 3.83LYY73 pKa = 10.74YY74 pKa = 10.93GDD76 pKa = 4.06EE77 pKa = 3.55WWGFRR82 pKa = 11.84MEE84 pKa = 4.38GGRR87 pKa = 11.84LIFPTGDD94 pKa = 2.67RR95 pKa = 11.84VAPQQLLAAIAILQIQAPDD114 pKa = 3.52DD115 pKa = 3.8AMTRR119 pKa = 11.84SKK121 pKa = 10.76LLKK124 pKa = 9.49YY125 pKa = 9.56ARR127 pKa = 11.84AMARR131 pKa = 11.84IKK133 pKa = 10.59GIKK136 pKa = 8.89
Molecular weight: 15.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1936 |
29 |
493 |
176.0 |
19.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.076 ± 0.877 | 1.963 ± 0.479 |
5.733 ± 0.848 | 5.01 ± 0.566 |
5.01 ± 0.327 | 6.198 ± 0.494 |
1.55 ± 0.399 | 8.368 ± 0.982 |
6.25 ± 0.559 | 8.626 ± 1.008 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.738 ± 0.422 | 4.545 ± 0.769 |
5.475 ± 0.71 | 3.564 ± 0.382 |
4.184 ± 0.864 | 6.353 ± 0.83 |
5.94 ± 0.669 | 5.785 ± 0.549 |
1.446 ± 0.407 | 4.184 ± 0.778 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
