
Pseudomonas sp. M47T1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; unclassified Pseudomonas
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
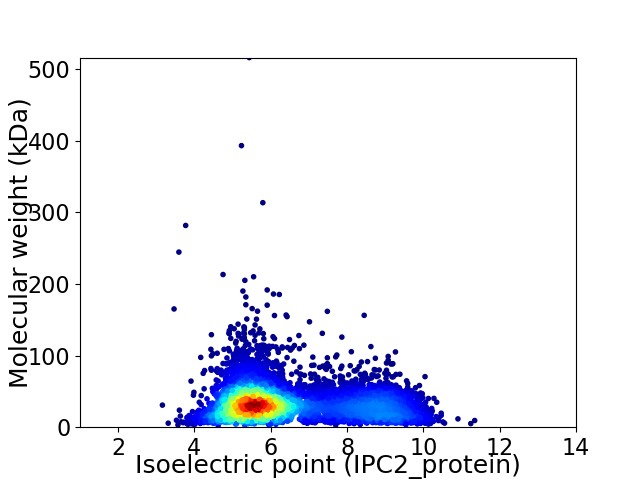
Virtual 2D-PAGE plot for 5675 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4N9F8|I4N9F8_9PSED Pyruvate kinase OS=Pseudomonas sp. M47T1 OX=1179778 GN=PMM47T1_02494 PE=3 SV=1
MM1 pKa = 7.23ATLIGVVSQVIGHH14 pKa = 5.69VFAMGADD21 pKa = 3.62GTRR24 pKa = 11.84RR25 pKa = 11.84EE26 pKa = 4.15VHH28 pKa = 5.55QGDD31 pKa = 3.68RR32 pKa = 11.84LFVGEE37 pKa = 4.4QLDD40 pKa = 3.89TGADD44 pKa = 3.49GAVAVHH50 pKa = 6.41LEE52 pKa = 3.95NGKK55 pKa = 9.12EE56 pKa = 3.87LTLGRR61 pKa = 11.84GSEE64 pKa = 4.35VTLSQQMLGGDD75 pKa = 3.97WAPHH79 pKa = 5.34VATQEE84 pKa = 3.92ATAPTQADD92 pKa = 3.65LTDD95 pKa = 3.48VQKK98 pKa = 10.66IQQAIAAGADD108 pKa = 3.37PTKK111 pKa = 10.6AAEE114 pKa = 4.09ATAAGPAALGTSTQGGAEE132 pKa = 4.25GGGHH136 pKa = 5.93SFVLLTEE143 pKa = 4.19TGGAIAPVLGYY154 pKa = 8.98ATAGFNGVADD164 pKa = 3.8FTVPQVAALTNANTAALVVDD184 pKa = 4.08NGVTLTGLGNGVSVDD199 pKa = 3.81EE200 pKa = 5.15ANLADD205 pKa = 4.15GSNPQTAALTQSSSFTVTAADD226 pKa = 3.77GLSNLVVGGITVVNNGAVVGIGQSITTGLGNTLTITGYY264 pKa = 10.92NPTTGQVSYY273 pKa = 10.66SYY275 pKa = 10.06TLTGPDD281 pKa = 3.51TQPAGGGANTVTEE294 pKa = 4.14NLGVSATDD302 pKa = 3.53SNGTNSSGSFTVTVVDD318 pKa = 5.09DD319 pKa = 4.13VPHH322 pKa = 7.42AINDD326 pKa = 3.83NPTTPATEE334 pKa = 3.99QNLVLTGNVLTNDD347 pKa = 2.84IQGADD352 pKa = 4.77RR353 pKa = 11.84IPITATSGPITAGTYY368 pKa = 8.03TGTYY372 pKa = 6.92GTLVLASDD380 pKa = 4.11GTYY383 pKa = 10.16TYY385 pKa = 9.95TLNAAGQAFNDD396 pKa = 3.51LHH398 pKa = 7.78GGGKK402 pKa = 8.49GTDD405 pKa = 3.11SFTYY409 pKa = 9.54TLNDD413 pKa = 3.62ADD415 pKa = 5.02GDD417 pKa = 3.93TSTAVLTLNINNINDD432 pKa = 3.74AVTITAPDD440 pKa = 3.58VNGVNTTVNEE450 pKa = 3.94ANLANGSSPAAAALTQTGQFTVSAPDD476 pKa = 3.5GLQTLTIGGVSVVTNGVGISSPVSVVTQLGTLTINSYY513 pKa = 10.74NATTGVVSYY522 pKa = 10.85SYY524 pKa = 10.12TLTHH528 pKa = 7.25ADD530 pKa = 3.3THH532 pKa = 6.94PDD534 pKa = 3.18VQGPNTITDD543 pKa = 4.03TFAVVATDD551 pKa = 3.64TDD553 pKa = 3.74GSTASTQLTTTIVDD567 pKa = 4.0DD568 pKa = 4.1VPKK571 pKa = 10.71AVADD575 pKa = 4.01TEE577 pKa = 4.69TVTEE581 pKa = 4.42GVTLQGNVLTNDD593 pKa = 2.95ASGADD598 pKa = 3.59GFGGVVGVKK607 pKa = 10.19AGSDD611 pKa = 3.01TSTAVSGGVGVAIEE625 pKa = 4.31GTYY628 pKa = 9.12GTLTINADD636 pKa = 3.51GSSSYY641 pKa = 10.19HH642 pKa = 6.03ANANITKK649 pKa = 9.94DD650 pKa = 3.45GAVSDD655 pKa = 3.86TFTYY659 pKa = 10.69SVADD663 pKa = 3.57KK664 pKa = 11.05DD665 pKa = 4.19GDD667 pKa = 3.54ISTTTVTVTINDD679 pKa = 3.89AGLKK683 pKa = 8.03ATNVDD688 pKa = 3.19GATVFEE694 pKa = 4.52AALPTGSHH702 pKa = 7.21PDD704 pKa = 3.04STAEE708 pKa = 3.97VGTGTVAGSVTGGVGALTYY727 pKa = 10.67SLQGADD733 pKa = 3.16ANGNVTSQYY742 pKa = 9.29GTLHH746 pKa = 6.71LNTDD750 pKa = 3.18GSYY753 pKa = 9.97TYY755 pKa = 9.64TLNVAPQVPGLGTLDD770 pKa = 3.45TFTLVTTDD778 pKa = 3.11SVGNQVDD785 pKa = 3.75SQVTIKK791 pKa = 10.53IVDD794 pKa = 3.84DD795 pKa = 3.97TPTAVPDD802 pKa = 3.83AVTMAEE808 pKa = 4.09GQIAQGNVLANDD820 pKa = 3.28ISGADD825 pKa = 3.44GFAGVVGVRR834 pKa = 11.84AGSDD838 pKa = 2.72TSTPVSGGVGTVIAGAYY855 pKa = 8.27GQLLIQADD863 pKa = 4.59GSAVYY868 pKa = 9.42HH869 pKa = 6.51ANANSVGEE877 pKa = 4.27AGATDD882 pKa = 3.78TFTYY886 pKa = 10.96SVMDD890 pKa = 3.67KK891 pKa = 10.99DD892 pKa = 4.55GDD894 pKa = 3.61ISTTTVTITINDD906 pKa = 3.71AGLVAKK912 pKa = 10.41NVDD915 pKa = 2.99GVTVFEE921 pKa = 4.74AALDD925 pKa = 3.89TTKK928 pKa = 11.11DD929 pKa = 3.83GNDD932 pKa = 3.34LAAGTSTGSHH942 pKa = 7.1PDD944 pKa = 3.29SPLEE948 pKa = 4.1TASGTVVGSVTGGVGTLTYY967 pKa = 10.45SVYY970 pKa = 10.9GSDD973 pKa = 3.7TNGDD977 pKa = 3.21MKK979 pKa = 10.94GQYY982 pKa = 10.39GVIHH986 pKa = 6.31MNADD990 pKa = 2.99GTYY993 pKa = 9.09TYY995 pKa = 10.53TLVTAPQVAGIGTTEE1010 pKa = 4.1NFVLVTTDD1018 pKa = 2.72SAGNHH1023 pKa = 4.97VVSDD1027 pKa = 3.69LTIKK1031 pKa = 10.19IVNDD1035 pKa = 3.38QPVALADD1042 pKa = 3.78SVTLTEE1048 pKa = 4.3GQVAQGNVLANDD1060 pKa = 3.34VSGADD1065 pKa = 3.41GFAGVVGVKK1074 pKa = 10.21AGSDD1078 pKa = 3.19TTTPVSGGVGSVVVGTYY1095 pKa = 10.87GEE1097 pKa = 4.32LVILADD1103 pKa = 3.98GSSIYY1108 pKa = 9.86HH1109 pKa = 6.54ANANSVGVTGATDD1122 pKa = 3.16VFTYY1126 pKa = 10.7SVADD1130 pKa = 3.55KK1131 pKa = 11.05DD1132 pKa = 4.19GDD1134 pKa = 3.54ISTTTVTININDD1146 pKa = 3.77SGLVAKK1152 pKa = 10.03NVNGVTVFEE1161 pKa = 4.48AALDD1165 pKa = 3.96LVKK1168 pKa = 10.83DD1169 pKa = 4.34GNDD1172 pKa = 3.54LAPGTSVGSHH1182 pKa = 7.15PDD1184 pKa = 3.1WTSEE1188 pKa = 4.12TASGTVAGSVTGGVGALTYY1207 pKa = 10.9SVFGSDD1213 pKa = 3.52ASGDD1217 pKa = 3.26IKK1219 pKa = 10.97GQYY1222 pKa = 9.49GVLHH1226 pKa = 6.16MNADD1230 pKa = 3.28GTYY1233 pKa = 9.17TYY1235 pKa = 10.5TLTTAPQVPGIGTTEE1250 pKa = 4.17SFTLVTTDD1258 pKa = 2.92SVGNHH1263 pKa = 5.22VASTLTVKK1271 pKa = 10.41IVDD1274 pKa = 4.27DD1275 pKa = 4.0QPHH1278 pKa = 6.97AFPDD1282 pKa = 4.33FAHH1285 pKa = 6.46LVEE1288 pKa = 4.8GQTAQGNVLANDD1300 pKa = 3.34VSGADD1305 pKa = 3.41GFAGVVGVKK1314 pKa = 10.47AGIDD1318 pKa = 3.38VSKK1321 pKa = 11.09AVIGGVGTVIQGQYY1335 pKa = 10.93GEE1337 pKa = 5.26LIIAADD1343 pKa = 3.82GTSIYY1348 pKa = 9.66HH1349 pKa = 6.67ANANSVGHH1357 pKa = 6.1FGIDD1361 pKa = 2.97VFTYY1365 pKa = 10.63SVADD1369 pKa = 3.55KK1370 pKa = 11.05DD1371 pKa = 4.19GDD1373 pKa = 3.53ISTTTLTIKK1382 pKa = 10.26IDD1384 pKa = 3.93DD1385 pKa = 4.39SGLKK1389 pKa = 10.05ASNVNGVTVFEE1400 pKa = 4.48AALDD1404 pKa = 3.96LVKK1407 pKa = 10.97DD1408 pKa = 4.28GNDD1411 pKa = 3.14LAAGNSVGSHH1421 pKa = 7.02PDD1423 pKa = 2.97WTSEE1427 pKa = 4.12TASGTVAGSVTGGVGALNYY1446 pKa = 10.29SVYY1449 pKa = 9.92GTDD1452 pKa = 3.2ASGDD1456 pKa = 3.22IKK1458 pKa = 10.95GQYY1461 pKa = 9.49GVLHH1465 pKa = 6.16MNADD1469 pKa = 3.28GTYY1472 pKa = 9.41TYY1474 pKa = 10.03TLTIAPQVAGIGTTEE1489 pKa = 4.04TFAVVTTDD1497 pKa = 3.04SVGNHH1502 pKa = 5.27VASTLTIKK1510 pKa = 10.47IVDD1513 pKa = 4.61DD1514 pKa = 3.73QPQAYY1519 pKa = 8.99PDD1521 pKa = 3.84FAHH1524 pKa = 6.08ITEE1527 pKa = 4.59GQTAQGNVLANDD1539 pKa = 3.34VSGADD1544 pKa = 3.41GFAGVVGVKK1553 pKa = 10.32AGADD1557 pKa = 3.3TSKK1560 pKa = 11.19AVTTGVGTVIQGQYY1574 pKa = 11.14GEE1576 pKa = 4.95LFIAADD1582 pKa = 3.91GTSTYY1587 pKa = 9.9HH1588 pKa = 7.16ANANSVNHH1596 pKa = 6.04VGIDD1600 pKa = 3.3VFTYY1604 pKa = 10.64SVADD1608 pKa = 3.55KK1609 pKa = 11.05DD1610 pKa = 4.19GDD1612 pKa = 3.53ISTTTLTIKK1621 pKa = 10.75VGDD1624 pKa = 3.84SHH1626 pKa = 6.79LTATNVDD1633 pKa = 3.36GVTVFEE1639 pKa = 4.6AALDD1643 pKa = 3.96LVKK1646 pKa = 10.97DD1647 pKa = 4.28GNDD1650 pKa = 3.14LAAGNSVGSHH1660 pKa = 7.02PDD1662 pKa = 2.97WTSEE1666 pKa = 4.12TASGTVAGSVTGGVGALNYY1685 pKa = 10.29SVYY1688 pKa = 9.92GTDD1691 pKa = 3.2ASGDD1695 pKa = 3.22IKK1697 pKa = 10.95GQYY1700 pKa = 9.49GVLHH1704 pKa = 6.16MNADD1708 pKa = 3.28GTYY1711 pKa = 9.17TYY1713 pKa = 10.5TLTTAPQVAGIGTTEE1728 pKa = 4.04TFAVVTTDD1736 pKa = 3.04SVGNHH1741 pKa = 5.59VASQLTIKK1749 pKa = 10.44IVDD1752 pKa = 4.51DD1753 pKa = 3.73QPQAYY1758 pKa = 8.1PDD1760 pKa = 3.59SAHH1763 pKa = 6.62ILEE1766 pKa = 4.8GQLAQGNVLQNDD1778 pKa = 3.38VSGADD1783 pKa = 3.38GFAGVVGVKK1792 pKa = 10.25AGSDD1796 pKa = 3.0TSTNVLGGVGTIIQGQYY1813 pKa = 8.53GQLVIAADD1821 pKa = 3.99GSSYY1825 pKa = 11.2YY1826 pKa = 10.11HH1827 pKa = 7.05ANPNSAAAGDD1837 pKa = 3.84VTDD1840 pKa = 3.6TFVYY1844 pKa = 10.51SVADD1848 pKa = 3.42KK1849 pKa = 11.24DD1850 pKa = 4.16GDD1852 pKa = 3.82VSTTTLTISIGNAGLHH1868 pKa = 5.77IVPGDD1873 pKa = 3.66GAGVTVAEE1881 pKa = 4.7AALDD1885 pKa = 3.98LNQDD1889 pKa = 3.84GKK1891 pKa = 10.87DD1892 pKa = 3.51LAAGTVTGSNPDD1904 pKa = 3.22STAEE1908 pKa = 4.03TGSGSVAGSVAGGVGTITYY1927 pKa = 10.25SIDD1930 pKa = 3.31GASNGLFTTAHH1941 pKa = 6.39GVIQLNADD1949 pKa = 3.37GSYY1952 pKa = 10.15TYY1954 pKa = 10.59TLTSAPKK1961 pKa = 9.5TDD1963 pKa = 3.75PSANNGADD1971 pKa = 3.34TTTDD1975 pKa = 3.04TVTYY1979 pKa = 10.19RR1980 pKa = 11.84ATDD1983 pKa = 3.45SQGNTITNQLTINITDD1999 pKa = 4.32DD2000 pKa = 3.56VPQAGAIDD2008 pKa = 3.71RR2009 pKa = 11.84AVIAPEE2015 pKa = 3.96VNSNVLLVIDD2025 pKa = 4.9ISGSMDD2031 pKa = 3.54DD2032 pKa = 3.76PSGVGKK2038 pKa = 10.83LSRR2041 pKa = 11.84LDD2043 pKa = 3.64LAKK2046 pKa = 10.94VSIDD2050 pKa = 3.71KK2051 pKa = 11.13LLDD2054 pKa = 3.3QYY2056 pKa = 12.06SGMGDD2061 pKa = 3.2VKK2063 pKa = 10.83VQVVTFSNGVSTPSSVWVDD2082 pKa = 3.13VATAKK2087 pKa = 10.69AYY2089 pKa = 9.85IAGLHH2094 pKa = 6.29ANGGTNYY2101 pKa = 10.75DD2102 pKa = 3.82YY2103 pKa = 11.32ALSGAEE2109 pKa = 4.31SAFNTNGKK2117 pKa = 9.31LVGGQNVAYY2126 pKa = 9.53FFSDD2130 pKa = 3.75GNPTLSSTHH2139 pKa = 5.74PTSGGSQSGSEE2150 pKa = 4.23TNPNLGDD2157 pKa = 4.53GIDD2160 pKa = 3.76TTEE2163 pKa = 3.91EE2164 pKa = 4.08KK2165 pKa = 10.76AWTDD2169 pKa = 3.47FLNANDD2175 pKa = 3.42IKK2177 pKa = 11.02AYY2179 pKa = 10.36AIGLGNGVDD2188 pKa = 3.78SQYY2191 pKa = 11.14LNPIAYY2197 pKa = 9.79DD2198 pKa = 3.48GTTGTNTNATVVTNLNNLGSVLSGTVQAVALTGSLLTNGTFGADD2242 pKa = 2.84GGFIKK2247 pKa = 10.63SITIDD2252 pKa = 3.52GVTYY2256 pKa = 9.67TYY2258 pKa = 10.88DD2259 pKa = 3.31PKK2261 pKa = 11.5ANNGEE2266 pKa = 4.18GGMTTAGGTSAGTFDD2281 pKa = 3.59TTTNSISVATSHH2293 pKa = 6.5GSTVVVDD2300 pKa = 4.3MDD2302 pKa = 3.74NGAYY2306 pKa = 8.61TYY2308 pKa = 11.03SSASSGSTATFTDD2321 pKa = 3.15KK2322 pKa = 10.54VTFVLADD2329 pKa = 3.48NDD2331 pKa = 4.07GDD2333 pKa = 4.15LSNAANLDD2341 pKa = 3.32ITVTPNSPPVAVADD2355 pKa = 3.75HH2356 pKa = 6.8VITNILSSTIIVPSEE2371 pKa = 3.4ALAANDD2377 pKa = 3.49TDD2379 pKa = 5.17ANGDD2383 pKa = 4.03PLTVANTGFDD2393 pKa = 3.57TGWAPKK2399 pKa = 10.42GADD2402 pKa = 3.52FTATTVQTIQFNGTSNLSSNQIKK2425 pKa = 9.73TIDD2428 pKa = 3.36RR2429 pKa = 11.84SDD2431 pKa = 3.87FNANTSNMTALLVVSGFLGAVGFGSNANASDD2462 pKa = 4.34SITVHH2467 pKa = 6.34LKK2469 pKa = 10.66AGEE2472 pKa = 4.13SLNLSDD2478 pKa = 5.67ASNSHH2483 pKa = 7.2LNMTWQLGNGSATTLGDD2500 pKa = 3.57GGVLTASTDD2509 pKa = 3.07GDD2511 pKa = 4.0YY2512 pKa = 10.78TIHH2515 pKa = 6.53VSHH2518 pKa = 7.2AANHH2522 pKa = 6.19TGQTAYY2528 pKa = 9.78TLDD2531 pKa = 3.34MSINYY2536 pKa = 9.6AGAANYY2542 pKa = 9.17TPDD2545 pKa = 3.92LPATYY2550 pKa = 9.37TVTDD2554 pKa = 3.42NHH2556 pKa = 6.63GGSDD2560 pKa = 3.3TAAATITYY2568 pKa = 10.04QDD2570 pKa = 3.27GHH2572 pKa = 6.13TLTGTAGDD2580 pKa = 5.47DD2581 pKa = 3.29ILISNDD2587 pKa = 3.22SGSTLLGLAGNDD2599 pKa = 3.52FMIGGAGNDD2608 pKa = 3.75KK2609 pKa = 10.7FDD2611 pKa = 4.1GGTGVNTVSYY2621 pKa = 9.4HH2622 pKa = 5.29AAKK2625 pKa = 10.72GPVTVDD2631 pKa = 3.32LSNTGVQDD2639 pKa = 3.66TVNAGHH2645 pKa = 7.03DD2646 pKa = 3.46QLINIQNLIGSDD2658 pKa = 3.82YY2659 pKa = 11.22GDD2661 pKa = 3.95HH2662 pKa = 6.56LTGNSQANVITGGAGNDD2679 pKa = 3.74VINGGGGNDD2688 pKa = 3.93TIIGGLGDD2696 pKa = 3.6NTLTGGDD2703 pKa = 3.54GADD2706 pKa = 3.33TFKK2709 pKa = 11.32YY2710 pKa = 10.12LAGNTGHH2717 pKa = 7.21DD2718 pKa = 4.14TITDD2722 pKa = 4.38FALGTDD2728 pKa = 4.08KK2729 pKa = 11.39LDD2731 pKa = 4.86LSQLLQGDD2739 pKa = 4.35GATASTLDD2747 pKa = 4.1DD2748 pKa = 3.64YY2749 pKa = 12.08LHH2751 pKa = 6.65FKK2753 pKa = 8.75VTGSGSGLVSSIDD2766 pKa = 3.47VSATAGGATTQTIDD2780 pKa = 3.5LQGVNVATHH2789 pKa = 5.56YY2790 pKa = 10.97GITPGAGGTISSHH2803 pKa = 5.95DD2804 pKa = 3.49TATIIGGMLGDD2815 pKa = 4.47HH2816 pKa = 5.92SLKK2819 pKa = 10.58VDD2821 pKa = 3.47VAA2823 pKa = 4.09
MM1 pKa = 7.23ATLIGVVSQVIGHH14 pKa = 5.69VFAMGADD21 pKa = 3.62GTRR24 pKa = 11.84RR25 pKa = 11.84EE26 pKa = 4.15VHH28 pKa = 5.55QGDD31 pKa = 3.68RR32 pKa = 11.84LFVGEE37 pKa = 4.4QLDD40 pKa = 3.89TGADD44 pKa = 3.49GAVAVHH50 pKa = 6.41LEE52 pKa = 3.95NGKK55 pKa = 9.12EE56 pKa = 3.87LTLGRR61 pKa = 11.84GSEE64 pKa = 4.35VTLSQQMLGGDD75 pKa = 3.97WAPHH79 pKa = 5.34VATQEE84 pKa = 3.92ATAPTQADD92 pKa = 3.65LTDD95 pKa = 3.48VQKK98 pKa = 10.66IQQAIAAGADD108 pKa = 3.37PTKK111 pKa = 10.6AAEE114 pKa = 4.09ATAAGPAALGTSTQGGAEE132 pKa = 4.25GGGHH136 pKa = 5.93SFVLLTEE143 pKa = 4.19TGGAIAPVLGYY154 pKa = 8.98ATAGFNGVADD164 pKa = 3.8FTVPQVAALTNANTAALVVDD184 pKa = 4.08NGVTLTGLGNGVSVDD199 pKa = 3.81EE200 pKa = 5.15ANLADD205 pKa = 4.15GSNPQTAALTQSSSFTVTAADD226 pKa = 3.77GLSNLVVGGITVVNNGAVVGIGQSITTGLGNTLTITGYY264 pKa = 10.92NPTTGQVSYY273 pKa = 10.66SYY275 pKa = 10.06TLTGPDD281 pKa = 3.51TQPAGGGANTVTEE294 pKa = 4.14NLGVSATDD302 pKa = 3.53SNGTNSSGSFTVTVVDD318 pKa = 5.09DD319 pKa = 4.13VPHH322 pKa = 7.42AINDD326 pKa = 3.83NPTTPATEE334 pKa = 3.99QNLVLTGNVLTNDD347 pKa = 2.84IQGADD352 pKa = 4.77RR353 pKa = 11.84IPITATSGPITAGTYY368 pKa = 8.03TGTYY372 pKa = 6.92GTLVLASDD380 pKa = 4.11GTYY383 pKa = 10.16TYY385 pKa = 9.95TLNAAGQAFNDD396 pKa = 3.51LHH398 pKa = 7.78GGGKK402 pKa = 8.49GTDD405 pKa = 3.11SFTYY409 pKa = 9.54TLNDD413 pKa = 3.62ADD415 pKa = 5.02GDD417 pKa = 3.93TSTAVLTLNINNINDD432 pKa = 3.74AVTITAPDD440 pKa = 3.58VNGVNTTVNEE450 pKa = 3.94ANLANGSSPAAAALTQTGQFTVSAPDD476 pKa = 3.5GLQTLTIGGVSVVTNGVGISSPVSVVTQLGTLTINSYY513 pKa = 10.74NATTGVVSYY522 pKa = 10.85SYY524 pKa = 10.12TLTHH528 pKa = 7.25ADD530 pKa = 3.3THH532 pKa = 6.94PDD534 pKa = 3.18VQGPNTITDD543 pKa = 4.03TFAVVATDD551 pKa = 3.64TDD553 pKa = 3.74GSTASTQLTTTIVDD567 pKa = 4.0DD568 pKa = 4.1VPKK571 pKa = 10.71AVADD575 pKa = 4.01TEE577 pKa = 4.69TVTEE581 pKa = 4.42GVTLQGNVLTNDD593 pKa = 2.95ASGADD598 pKa = 3.59GFGGVVGVKK607 pKa = 10.19AGSDD611 pKa = 3.01TSTAVSGGVGVAIEE625 pKa = 4.31GTYY628 pKa = 9.12GTLTINADD636 pKa = 3.51GSSSYY641 pKa = 10.19HH642 pKa = 6.03ANANITKK649 pKa = 9.94DD650 pKa = 3.45GAVSDD655 pKa = 3.86TFTYY659 pKa = 10.69SVADD663 pKa = 3.57KK664 pKa = 11.05DD665 pKa = 4.19GDD667 pKa = 3.54ISTTTVTVTINDD679 pKa = 3.89AGLKK683 pKa = 8.03ATNVDD688 pKa = 3.19GATVFEE694 pKa = 4.52AALPTGSHH702 pKa = 7.21PDD704 pKa = 3.04STAEE708 pKa = 3.97VGTGTVAGSVTGGVGALTYY727 pKa = 10.67SLQGADD733 pKa = 3.16ANGNVTSQYY742 pKa = 9.29GTLHH746 pKa = 6.71LNTDD750 pKa = 3.18GSYY753 pKa = 9.97TYY755 pKa = 9.64TLNVAPQVPGLGTLDD770 pKa = 3.45TFTLVTTDD778 pKa = 3.11SVGNQVDD785 pKa = 3.75SQVTIKK791 pKa = 10.53IVDD794 pKa = 3.84DD795 pKa = 3.97TPTAVPDD802 pKa = 3.83AVTMAEE808 pKa = 4.09GQIAQGNVLANDD820 pKa = 3.28ISGADD825 pKa = 3.44GFAGVVGVRR834 pKa = 11.84AGSDD838 pKa = 2.72TSTPVSGGVGTVIAGAYY855 pKa = 8.27GQLLIQADD863 pKa = 4.59GSAVYY868 pKa = 9.42HH869 pKa = 6.51ANANSVGEE877 pKa = 4.27AGATDD882 pKa = 3.78TFTYY886 pKa = 10.96SVMDD890 pKa = 3.67KK891 pKa = 10.99DD892 pKa = 4.55GDD894 pKa = 3.61ISTTTVTITINDD906 pKa = 3.71AGLVAKK912 pKa = 10.41NVDD915 pKa = 2.99GVTVFEE921 pKa = 4.74AALDD925 pKa = 3.89TTKK928 pKa = 11.11DD929 pKa = 3.83GNDD932 pKa = 3.34LAAGTSTGSHH942 pKa = 7.1PDD944 pKa = 3.29SPLEE948 pKa = 4.1TASGTVVGSVTGGVGTLTYY967 pKa = 10.45SVYY970 pKa = 10.9GSDD973 pKa = 3.7TNGDD977 pKa = 3.21MKK979 pKa = 10.94GQYY982 pKa = 10.39GVIHH986 pKa = 6.31MNADD990 pKa = 2.99GTYY993 pKa = 9.09TYY995 pKa = 10.53TLVTAPQVAGIGTTEE1010 pKa = 4.1NFVLVTTDD1018 pKa = 2.72SAGNHH1023 pKa = 4.97VVSDD1027 pKa = 3.69LTIKK1031 pKa = 10.19IVNDD1035 pKa = 3.38QPVALADD1042 pKa = 3.78SVTLTEE1048 pKa = 4.3GQVAQGNVLANDD1060 pKa = 3.34VSGADD1065 pKa = 3.41GFAGVVGVKK1074 pKa = 10.21AGSDD1078 pKa = 3.19TTTPVSGGVGSVVVGTYY1095 pKa = 10.87GEE1097 pKa = 4.32LVILADD1103 pKa = 3.98GSSIYY1108 pKa = 9.86HH1109 pKa = 6.54ANANSVGVTGATDD1122 pKa = 3.16VFTYY1126 pKa = 10.7SVADD1130 pKa = 3.55KK1131 pKa = 11.05DD1132 pKa = 4.19GDD1134 pKa = 3.54ISTTTVTININDD1146 pKa = 3.77SGLVAKK1152 pKa = 10.03NVNGVTVFEE1161 pKa = 4.48AALDD1165 pKa = 3.96LVKK1168 pKa = 10.83DD1169 pKa = 4.34GNDD1172 pKa = 3.54LAPGTSVGSHH1182 pKa = 7.15PDD1184 pKa = 3.1WTSEE1188 pKa = 4.12TASGTVAGSVTGGVGALTYY1207 pKa = 10.9SVFGSDD1213 pKa = 3.52ASGDD1217 pKa = 3.26IKK1219 pKa = 10.97GQYY1222 pKa = 9.49GVLHH1226 pKa = 6.16MNADD1230 pKa = 3.28GTYY1233 pKa = 9.17TYY1235 pKa = 10.5TLTTAPQVPGIGTTEE1250 pKa = 4.17SFTLVTTDD1258 pKa = 2.92SVGNHH1263 pKa = 5.22VASTLTVKK1271 pKa = 10.41IVDD1274 pKa = 4.27DD1275 pKa = 4.0QPHH1278 pKa = 6.97AFPDD1282 pKa = 4.33FAHH1285 pKa = 6.46LVEE1288 pKa = 4.8GQTAQGNVLANDD1300 pKa = 3.34VSGADD1305 pKa = 3.41GFAGVVGVKK1314 pKa = 10.47AGIDD1318 pKa = 3.38VSKK1321 pKa = 11.09AVIGGVGTVIQGQYY1335 pKa = 10.93GEE1337 pKa = 5.26LIIAADD1343 pKa = 3.82GTSIYY1348 pKa = 9.66HH1349 pKa = 6.67ANANSVGHH1357 pKa = 6.1FGIDD1361 pKa = 2.97VFTYY1365 pKa = 10.63SVADD1369 pKa = 3.55KK1370 pKa = 11.05DD1371 pKa = 4.19GDD1373 pKa = 3.53ISTTTLTIKK1382 pKa = 10.26IDD1384 pKa = 3.93DD1385 pKa = 4.39SGLKK1389 pKa = 10.05ASNVNGVTVFEE1400 pKa = 4.48AALDD1404 pKa = 3.96LVKK1407 pKa = 10.97DD1408 pKa = 4.28GNDD1411 pKa = 3.14LAAGNSVGSHH1421 pKa = 7.02PDD1423 pKa = 2.97WTSEE1427 pKa = 4.12TASGTVAGSVTGGVGALNYY1446 pKa = 10.29SVYY1449 pKa = 9.92GTDD1452 pKa = 3.2ASGDD1456 pKa = 3.22IKK1458 pKa = 10.95GQYY1461 pKa = 9.49GVLHH1465 pKa = 6.16MNADD1469 pKa = 3.28GTYY1472 pKa = 9.41TYY1474 pKa = 10.03TLTIAPQVAGIGTTEE1489 pKa = 4.04TFAVVTTDD1497 pKa = 3.04SVGNHH1502 pKa = 5.27VASTLTIKK1510 pKa = 10.47IVDD1513 pKa = 4.61DD1514 pKa = 3.73QPQAYY1519 pKa = 8.99PDD1521 pKa = 3.84FAHH1524 pKa = 6.08ITEE1527 pKa = 4.59GQTAQGNVLANDD1539 pKa = 3.34VSGADD1544 pKa = 3.41GFAGVVGVKK1553 pKa = 10.32AGADD1557 pKa = 3.3TSKK1560 pKa = 11.19AVTTGVGTVIQGQYY1574 pKa = 11.14GEE1576 pKa = 4.95LFIAADD1582 pKa = 3.91GTSTYY1587 pKa = 9.9HH1588 pKa = 7.16ANANSVNHH1596 pKa = 6.04VGIDD1600 pKa = 3.3VFTYY1604 pKa = 10.64SVADD1608 pKa = 3.55KK1609 pKa = 11.05DD1610 pKa = 4.19GDD1612 pKa = 3.53ISTTTLTIKK1621 pKa = 10.75VGDD1624 pKa = 3.84SHH1626 pKa = 6.79LTATNVDD1633 pKa = 3.36GVTVFEE1639 pKa = 4.6AALDD1643 pKa = 3.96LVKK1646 pKa = 10.97DD1647 pKa = 4.28GNDD1650 pKa = 3.14LAAGNSVGSHH1660 pKa = 7.02PDD1662 pKa = 2.97WTSEE1666 pKa = 4.12TASGTVAGSVTGGVGALNYY1685 pKa = 10.29SVYY1688 pKa = 9.92GTDD1691 pKa = 3.2ASGDD1695 pKa = 3.22IKK1697 pKa = 10.95GQYY1700 pKa = 9.49GVLHH1704 pKa = 6.16MNADD1708 pKa = 3.28GTYY1711 pKa = 9.17TYY1713 pKa = 10.5TLTTAPQVAGIGTTEE1728 pKa = 4.04TFAVVTTDD1736 pKa = 3.04SVGNHH1741 pKa = 5.59VASQLTIKK1749 pKa = 10.44IVDD1752 pKa = 4.51DD1753 pKa = 3.73QPQAYY1758 pKa = 8.1PDD1760 pKa = 3.59SAHH1763 pKa = 6.62ILEE1766 pKa = 4.8GQLAQGNVLQNDD1778 pKa = 3.38VSGADD1783 pKa = 3.38GFAGVVGVKK1792 pKa = 10.25AGSDD1796 pKa = 3.0TSTNVLGGVGTIIQGQYY1813 pKa = 8.53GQLVIAADD1821 pKa = 3.99GSSYY1825 pKa = 11.2YY1826 pKa = 10.11HH1827 pKa = 7.05ANPNSAAAGDD1837 pKa = 3.84VTDD1840 pKa = 3.6TFVYY1844 pKa = 10.51SVADD1848 pKa = 3.42KK1849 pKa = 11.24DD1850 pKa = 4.16GDD1852 pKa = 3.82VSTTTLTISIGNAGLHH1868 pKa = 5.77IVPGDD1873 pKa = 3.66GAGVTVAEE1881 pKa = 4.7AALDD1885 pKa = 3.98LNQDD1889 pKa = 3.84GKK1891 pKa = 10.87DD1892 pKa = 3.51LAAGTVTGSNPDD1904 pKa = 3.22STAEE1908 pKa = 4.03TGSGSVAGSVAGGVGTITYY1927 pKa = 10.25SIDD1930 pKa = 3.31GASNGLFTTAHH1941 pKa = 6.39GVIQLNADD1949 pKa = 3.37GSYY1952 pKa = 10.15TYY1954 pKa = 10.59TLTSAPKK1961 pKa = 9.5TDD1963 pKa = 3.75PSANNGADD1971 pKa = 3.34TTTDD1975 pKa = 3.04TVTYY1979 pKa = 10.19RR1980 pKa = 11.84ATDD1983 pKa = 3.45SQGNTITNQLTINITDD1999 pKa = 4.32DD2000 pKa = 3.56VPQAGAIDD2008 pKa = 3.71RR2009 pKa = 11.84AVIAPEE2015 pKa = 3.96VNSNVLLVIDD2025 pKa = 4.9ISGSMDD2031 pKa = 3.54DD2032 pKa = 3.76PSGVGKK2038 pKa = 10.83LSRR2041 pKa = 11.84LDD2043 pKa = 3.64LAKK2046 pKa = 10.94VSIDD2050 pKa = 3.71KK2051 pKa = 11.13LLDD2054 pKa = 3.3QYY2056 pKa = 12.06SGMGDD2061 pKa = 3.2VKK2063 pKa = 10.83VQVVTFSNGVSTPSSVWVDD2082 pKa = 3.13VATAKK2087 pKa = 10.69AYY2089 pKa = 9.85IAGLHH2094 pKa = 6.29ANGGTNYY2101 pKa = 10.75DD2102 pKa = 3.82YY2103 pKa = 11.32ALSGAEE2109 pKa = 4.31SAFNTNGKK2117 pKa = 9.31LVGGQNVAYY2126 pKa = 9.53FFSDD2130 pKa = 3.75GNPTLSSTHH2139 pKa = 5.74PTSGGSQSGSEE2150 pKa = 4.23TNPNLGDD2157 pKa = 4.53GIDD2160 pKa = 3.76TTEE2163 pKa = 3.91EE2164 pKa = 4.08KK2165 pKa = 10.76AWTDD2169 pKa = 3.47FLNANDD2175 pKa = 3.42IKK2177 pKa = 11.02AYY2179 pKa = 10.36AIGLGNGVDD2188 pKa = 3.78SQYY2191 pKa = 11.14LNPIAYY2197 pKa = 9.79DD2198 pKa = 3.48GTTGTNTNATVVTNLNNLGSVLSGTVQAVALTGSLLTNGTFGADD2242 pKa = 2.84GGFIKK2247 pKa = 10.63SITIDD2252 pKa = 3.52GVTYY2256 pKa = 9.67TYY2258 pKa = 10.88DD2259 pKa = 3.31PKK2261 pKa = 11.5ANNGEE2266 pKa = 4.18GGMTTAGGTSAGTFDD2281 pKa = 3.59TTTNSISVATSHH2293 pKa = 6.5GSTVVVDD2300 pKa = 4.3MDD2302 pKa = 3.74NGAYY2306 pKa = 8.61TYY2308 pKa = 11.03SSASSGSTATFTDD2321 pKa = 3.15KK2322 pKa = 10.54VTFVLADD2329 pKa = 3.48NDD2331 pKa = 4.07GDD2333 pKa = 4.15LSNAANLDD2341 pKa = 3.32ITVTPNSPPVAVADD2355 pKa = 3.75HH2356 pKa = 6.8VITNILSSTIIVPSEE2371 pKa = 3.4ALAANDD2377 pKa = 3.49TDD2379 pKa = 5.17ANGDD2383 pKa = 4.03PLTVANTGFDD2393 pKa = 3.57TGWAPKK2399 pKa = 10.42GADD2402 pKa = 3.52FTATTVQTIQFNGTSNLSSNQIKK2425 pKa = 9.73TIDD2428 pKa = 3.36RR2429 pKa = 11.84SDD2431 pKa = 3.87FNANTSNMTALLVVSGFLGAVGFGSNANASDD2462 pKa = 4.34SITVHH2467 pKa = 6.34LKK2469 pKa = 10.66AGEE2472 pKa = 4.13SLNLSDD2478 pKa = 5.67ASNSHH2483 pKa = 7.2LNMTWQLGNGSATTLGDD2500 pKa = 3.57GGVLTASTDD2509 pKa = 3.07GDD2511 pKa = 4.0YY2512 pKa = 10.78TIHH2515 pKa = 6.53VSHH2518 pKa = 7.2AANHH2522 pKa = 6.19TGQTAYY2528 pKa = 9.78TLDD2531 pKa = 3.34MSINYY2536 pKa = 9.6AGAANYY2542 pKa = 9.17TPDD2545 pKa = 3.92LPATYY2550 pKa = 9.37TVTDD2554 pKa = 3.42NHH2556 pKa = 6.63GGSDD2560 pKa = 3.3TAAATITYY2568 pKa = 10.04QDD2570 pKa = 3.27GHH2572 pKa = 6.13TLTGTAGDD2580 pKa = 5.47DD2581 pKa = 3.29ILISNDD2587 pKa = 3.22SGSTLLGLAGNDD2599 pKa = 3.52FMIGGAGNDD2608 pKa = 3.75KK2609 pKa = 10.7FDD2611 pKa = 4.1GGTGVNTVSYY2621 pKa = 9.4HH2622 pKa = 5.29AAKK2625 pKa = 10.72GPVTVDD2631 pKa = 3.32LSNTGVQDD2639 pKa = 3.66TVNAGHH2645 pKa = 7.03DD2646 pKa = 3.46QLINIQNLIGSDD2658 pKa = 3.82YY2659 pKa = 11.22GDD2661 pKa = 3.95HH2662 pKa = 6.56LTGNSQANVITGGAGNDD2679 pKa = 3.74VINGGGGNDD2688 pKa = 3.93TIIGGLGDD2696 pKa = 3.6NTLTGGDD2703 pKa = 3.54GADD2706 pKa = 3.33TFKK2709 pKa = 11.32YY2710 pKa = 10.12LAGNTGHH2717 pKa = 7.21DD2718 pKa = 4.14TITDD2722 pKa = 4.38FALGTDD2728 pKa = 4.08KK2729 pKa = 11.39LDD2731 pKa = 4.86LSQLLQGDD2739 pKa = 4.35GATASTLDD2747 pKa = 4.1DD2748 pKa = 3.64YY2749 pKa = 12.08LHH2751 pKa = 6.65FKK2753 pKa = 8.75VTGSGSGLVSSIDD2766 pKa = 3.47VSATAGGATTQTIDD2780 pKa = 3.5LQGVNVATHH2789 pKa = 5.56YY2790 pKa = 10.97GITPGAGGTISSHH2803 pKa = 5.95DD2804 pKa = 3.49TATIIGGMLGDD2815 pKa = 4.47HH2816 pKa = 5.92SLKK2819 pKa = 10.58VDD2821 pKa = 3.47VAA2823 pKa = 4.09
Molecular weight: 281.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4N9M8|I4N9M8_9PSED Tricarboxylate transport protein TctC OS=Pseudomonas sp. M47T1 OX=1179778 GN=PMM47T1_02854 PE=3 SV=1
MM1 pKa = 7.79APRR4 pKa = 11.84TTTPVITFGRR14 pKa = 11.84RR15 pKa = 11.84ATTRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84LITTVLHH28 pKa = 6.23PAAVTGQAGRR38 pKa = 11.84AGTVIAHH45 pKa = 5.9RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84GAARR54 pKa = 11.84VAQAGATGRR63 pKa = 11.84GTATMTTMVALATAGLAAAAAKK85 pKa = 9.92AASKK89 pKa = 11.35ANLL92 pKa = 3.33
MM1 pKa = 7.79APRR4 pKa = 11.84TTTPVITFGRR14 pKa = 11.84RR15 pKa = 11.84ATTRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84LITTVLHH28 pKa = 6.23PAAVTGQAGRR38 pKa = 11.84AGTVIAHH45 pKa = 5.9RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84GAARR54 pKa = 11.84VAQAGATGRR63 pKa = 11.84GTATMTTMVALATAGLAAAAAKK85 pKa = 9.92AASKK89 pKa = 11.35ANLL92 pKa = 3.33
Molecular weight: 9.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1837506 |
22 |
4676 |
323.8 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.419 ± 0.045 | 0.988 ± 0.011 |
5.427 ± 0.024 | 5.2 ± 0.036 |
3.585 ± 0.023 | 8.076 ± 0.033 |
2.442 ± 0.017 | 4.594 ± 0.024 |
3.274 ± 0.029 | 11.69 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.344 ± 0.017 | 2.992 ± 0.02 |
4.949 ± 0.024 | 4.712 ± 0.027 |
6.288 ± 0.035 | 5.651 ± 0.028 |
5.036 ± 0.037 | 7.334 ± 0.027 |
1.466 ± 0.014 | 2.532 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
