
[Clostridium] methylpentosum DSM 5476
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillospiraceae incertae sedis; [Clostridium] methylpentosum
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
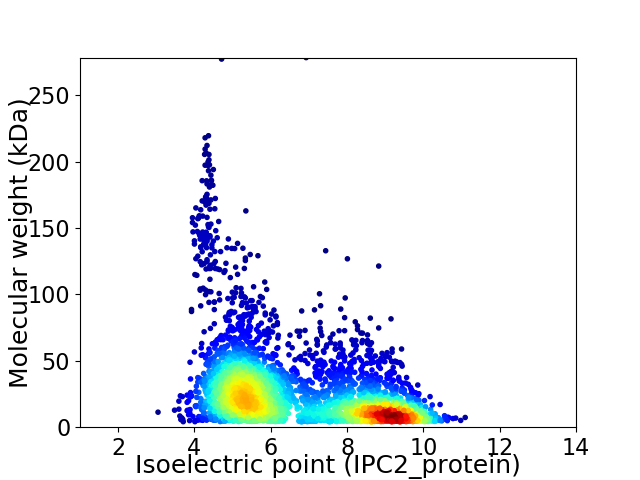
Virtual 2D-PAGE plot for 3907 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0EIG0|C0EIG0_9FIRM Spermidine/putrescine import ATP-binding protein PotA OS=[Clostridium] methylpentosum DSM 5476 OX=537013 GN=potA PE=3 SV=1
MM1 pKa = 7.41NEE3 pKa = 3.41TWEE6 pKa = 4.56DD7 pKa = 3.62VAGDD11 pKa = 4.13ITDD14 pKa = 2.58KK15 pKa = 10.77WYY17 pKa = 8.9ITNVCIKK24 pKa = 10.43GFTNPLPEE32 pKa = 5.44SDD34 pKa = 3.54TAVASVRR41 pKa = 11.84FSEE44 pKa = 4.47MEE46 pKa = 4.09GPLADD51 pKa = 4.01GTDD54 pKa = 3.1ISLTAEE60 pKa = 4.08GAEE63 pKa = 4.09EE64 pKa = 4.11IYY66 pKa = 11.13YY67 pKa = 10.7SVDD70 pKa = 2.52GAAYY74 pKa = 9.64QKK76 pKa = 9.46YY77 pKa = 6.42TAPLTLDD84 pKa = 4.31FAQQDD89 pKa = 3.65AHH91 pKa = 5.97TVAAYY96 pKa = 9.94AVDD99 pKa = 3.51NGKK102 pKa = 9.71QGNTVEE108 pKa = 4.17KK109 pKa = 10.38VYY111 pKa = 10.64TKK113 pKa = 10.87AVAQLTDD120 pKa = 3.65LALKK124 pKa = 9.48YY125 pKa = 10.65DD126 pKa = 4.08GVVEE130 pKa = 4.11HH131 pKa = 7.32CEE133 pKa = 3.85TDD135 pKa = 3.1GLTKK139 pKa = 10.62HH140 pKa = 6.25FVYY143 pKa = 10.71LPNCADD149 pKa = 3.49NVQVMAQSADD159 pKa = 3.49KK160 pKa = 10.84IRR162 pKa = 11.84VNGQMLNSSDD172 pKa = 3.22WSDD175 pKa = 4.26GISLTPGEE183 pKa = 4.37TTEE186 pKa = 3.81ITIEE190 pKa = 4.01VTGSGKK196 pKa = 8.37TPSTYY201 pKa = 9.73TLEE204 pKa = 4.83AYY206 pKa = 10.4RR207 pKa = 11.84SMLIFDD213 pKa = 4.51YY214 pKa = 10.77EE215 pKa = 4.47AEE217 pKa = 4.34TVSYY221 pKa = 11.24DD222 pKa = 3.36DD223 pKa = 3.4TNYY226 pKa = 10.52TVMDD230 pKa = 4.06TQQNEE235 pKa = 4.32IKK237 pKa = 10.7SGDD240 pKa = 3.89SIAALISTEE249 pKa = 4.2EE250 pKa = 4.12KK251 pKa = 10.32TEE253 pKa = 4.02LTVTPIAGGDD263 pKa = 3.52NLIEE267 pKa = 4.0YY268 pKa = 9.05VPKK271 pKa = 10.53RR272 pKa = 11.84ILVSPTGINYY282 pKa = 10.21ALEE285 pKa = 4.37EE286 pKa = 4.29TDD288 pKa = 4.86SSFSDD293 pKa = 4.18LYY295 pKa = 11.15AYY297 pKa = 9.79SANADD302 pKa = 3.2MSEE305 pKa = 3.95AVQCEE310 pKa = 4.09YY311 pKa = 11.29GQTIPLTPGVDD322 pKa = 3.56VYY324 pKa = 10.78IQRR327 pKa = 11.84QATDD331 pKa = 3.52RR332 pKa = 11.84DD333 pKa = 4.49FISAIYY339 pKa = 9.42HH340 pKa = 5.74LEE342 pKa = 4.05VPRR345 pKa = 11.84RR346 pKa = 11.84PAAPEE351 pKa = 3.73VTAEE355 pKa = 4.35EE356 pKa = 4.23ITDD359 pKa = 3.64TSVTLQAMEE368 pKa = 4.28GALYY372 pKa = 10.61SAGGDD377 pKa = 3.39WQDD380 pKa = 3.3TPVFTDD386 pKa = 3.64LTPGTEE392 pKa = 4.15YY393 pKa = 10.63TFQVCLMATEE403 pKa = 4.0NAFRR407 pKa = 11.84SEE409 pKa = 3.82YY410 pKa = 10.37GAAKK414 pKa = 8.73ITTKK418 pKa = 10.14IPQVNPSDD426 pKa = 3.56YY427 pKa = 10.48SFEE430 pKa = 4.12VKK432 pKa = 10.71YY433 pKa = 11.12VDD435 pKa = 4.67GEE437 pKa = 4.54GNPVPGGGIISFEE450 pKa = 3.88EE451 pKa = 3.61VGPYY455 pKa = 9.68NRR457 pKa = 11.84EE458 pKa = 4.47DD459 pKa = 3.17IPLPYY464 pKa = 10.21GYY466 pKa = 8.53MQIIPAHH473 pKa = 6.85PDD475 pKa = 3.31EE476 pKa = 5.74DD477 pKa = 4.06WLFPTALEE485 pKa = 4.3WVDD488 pKa = 4.72GEE490 pKa = 4.47WKK492 pKa = 8.52VTNPVVEE499 pKa = 4.12IMVEE503 pKa = 3.97KK504 pKa = 9.62MASVDD509 pKa = 4.19IIFKK513 pKa = 9.99TPDD516 pKa = 3.05GNILEE521 pKa = 4.53DD522 pKa = 3.41WNYY525 pKa = 8.18TRR527 pKa = 11.84YY528 pKa = 10.06YY529 pKa = 10.24DD530 pKa = 4.04SEE532 pKa = 5.16GGGIEE537 pKa = 4.21TVPAPEE543 pKa = 4.89GYY545 pKa = 10.48EE546 pKa = 3.75FVGEE550 pKa = 3.7NTYY553 pKa = 10.93AVEE556 pKa = 4.48VIRR559 pKa = 11.84DD560 pKa = 3.63EE561 pKa = 4.28TGKK564 pKa = 10.75LVADD568 pKa = 4.18PSEE571 pKa = 3.92VTFWVKK577 pKa = 10.58EE578 pKa = 3.91IGSEE582 pKa = 4.29IPVTPSDD589 pKa = 3.49YY590 pKa = 11.03SFEE593 pKa = 4.1VKK595 pKa = 10.87YY596 pKa = 10.59IDD598 pKa = 4.83GEE600 pKa = 4.55GNPVPGGGTISFEE613 pKa = 3.83EE614 pKa = 4.06AGPYY618 pKa = 9.36NRR620 pKa = 11.84EE621 pKa = 4.74DD622 pKa = 3.13IPLPYY627 pKa = 10.21GYY629 pKa = 8.53MQIIPAHH636 pKa = 6.85PDD638 pKa = 3.31EE639 pKa = 5.76DD640 pKa = 3.98WLFPTSLVLVDD651 pKa = 4.98GEE653 pKa = 4.47WKK655 pKa = 8.46VTNPVVEE662 pKa = 4.12IMVEE666 pKa = 3.97KK667 pKa = 9.62MASVDD672 pKa = 5.05IIFQTPDD679 pKa = 2.95GNILEE684 pKa = 4.53DD685 pKa = 3.41WNYY688 pKa = 8.18TRR690 pKa = 11.84YY691 pKa = 10.06YY692 pKa = 10.24DD693 pKa = 4.04SEE695 pKa = 5.16GGGIEE700 pKa = 4.21TVPAPEE706 pKa = 4.77GYY708 pKa = 10.55EE709 pKa = 3.95FIGEE713 pKa = 3.68NTYY716 pKa = 10.89AVEE719 pKa = 4.48VIRR722 pKa = 11.84DD723 pKa = 3.63EE724 pKa = 4.28TGKK727 pKa = 10.75LVADD731 pKa = 4.06PSEE734 pKa = 4.12VIFIVKK740 pKa = 10.42AIDD743 pKa = 3.4TEE745 pKa = 4.88GPSDD749 pKa = 4.01PEE751 pKa = 4.22EE752 pKa = 4.38PSTPGGTNTPEE763 pKa = 4.34GNSPQTGDD771 pKa = 4.34DD772 pKa = 3.45SHH774 pKa = 8.32ASNLLLLILVSAGTLLLVTIIRR796 pKa = 11.84NQKK799 pKa = 8.33KK800 pKa = 9.75KK801 pKa = 11.06EE802 pKa = 3.95MTTEE806 pKa = 3.89EE807 pKa = 4.12
MM1 pKa = 7.41NEE3 pKa = 3.41TWEE6 pKa = 4.56DD7 pKa = 3.62VAGDD11 pKa = 4.13ITDD14 pKa = 2.58KK15 pKa = 10.77WYY17 pKa = 8.9ITNVCIKK24 pKa = 10.43GFTNPLPEE32 pKa = 5.44SDD34 pKa = 3.54TAVASVRR41 pKa = 11.84FSEE44 pKa = 4.47MEE46 pKa = 4.09GPLADD51 pKa = 4.01GTDD54 pKa = 3.1ISLTAEE60 pKa = 4.08GAEE63 pKa = 4.09EE64 pKa = 4.11IYY66 pKa = 11.13YY67 pKa = 10.7SVDD70 pKa = 2.52GAAYY74 pKa = 9.64QKK76 pKa = 9.46YY77 pKa = 6.42TAPLTLDD84 pKa = 4.31FAQQDD89 pKa = 3.65AHH91 pKa = 5.97TVAAYY96 pKa = 9.94AVDD99 pKa = 3.51NGKK102 pKa = 9.71QGNTVEE108 pKa = 4.17KK109 pKa = 10.38VYY111 pKa = 10.64TKK113 pKa = 10.87AVAQLTDD120 pKa = 3.65LALKK124 pKa = 9.48YY125 pKa = 10.65DD126 pKa = 4.08GVVEE130 pKa = 4.11HH131 pKa = 7.32CEE133 pKa = 3.85TDD135 pKa = 3.1GLTKK139 pKa = 10.62HH140 pKa = 6.25FVYY143 pKa = 10.71LPNCADD149 pKa = 3.49NVQVMAQSADD159 pKa = 3.49KK160 pKa = 10.84IRR162 pKa = 11.84VNGQMLNSSDD172 pKa = 3.22WSDD175 pKa = 4.26GISLTPGEE183 pKa = 4.37TTEE186 pKa = 3.81ITIEE190 pKa = 4.01VTGSGKK196 pKa = 8.37TPSTYY201 pKa = 9.73TLEE204 pKa = 4.83AYY206 pKa = 10.4RR207 pKa = 11.84SMLIFDD213 pKa = 4.51YY214 pKa = 10.77EE215 pKa = 4.47AEE217 pKa = 4.34TVSYY221 pKa = 11.24DD222 pKa = 3.36DD223 pKa = 3.4TNYY226 pKa = 10.52TVMDD230 pKa = 4.06TQQNEE235 pKa = 4.32IKK237 pKa = 10.7SGDD240 pKa = 3.89SIAALISTEE249 pKa = 4.2EE250 pKa = 4.12KK251 pKa = 10.32TEE253 pKa = 4.02LTVTPIAGGDD263 pKa = 3.52NLIEE267 pKa = 4.0YY268 pKa = 9.05VPKK271 pKa = 10.53RR272 pKa = 11.84ILVSPTGINYY282 pKa = 10.21ALEE285 pKa = 4.37EE286 pKa = 4.29TDD288 pKa = 4.86SSFSDD293 pKa = 4.18LYY295 pKa = 11.15AYY297 pKa = 9.79SANADD302 pKa = 3.2MSEE305 pKa = 3.95AVQCEE310 pKa = 4.09YY311 pKa = 11.29GQTIPLTPGVDD322 pKa = 3.56VYY324 pKa = 10.78IQRR327 pKa = 11.84QATDD331 pKa = 3.52RR332 pKa = 11.84DD333 pKa = 4.49FISAIYY339 pKa = 9.42HH340 pKa = 5.74LEE342 pKa = 4.05VPRR345 pKa = 11.84RR346 pKa = 11.84PAAPEE351 pKa = 3.73VTAEE355 pKa = 4.35EE356 pKa = 4.23ITDD359 pKa = 3.64TSVTLQAMEE368 pKa = 4.28GALYY372 pKa = 10.61SAGGDD377 pKa = 3.39WQDD380 pKa = 3.3TPVFTDD386 pKa = 3.64LTPGTEE392 pKa = 4.15YY393 pKa = 10.63TFQVCLMATEE403 pKa = 4.0NAFRR407 pKa = 11.84SEE409 pKa = 3.82YY410 pKa = 10.37GAAKK414 pKa = 8.73ITTKK418 pKa = 10.14IPQVNPSDD426 pKa = 3.56YY427 pKa = 10.48SFEE430 pKa = 4.12VKK432 pKa = 10.71YY433 pKa = 11.12VDD435 pKa = 4.67GEE437 pKa = 4.54GNPVPGGGIISFEE450 pKa = 3.88EE451 pKa = 3.61VGPYY455 pKa = 9.68NRR457 pKa = 11.84EE458 pKa = 4.47DD459 pKa = 3.17IPLPYY464 pKa = 10.21GYY466 pKa = 8.53MQIIPAHH473 pKa = 6.85PDD475 pKa = 3.31EE476 pKa = 5.74DD477 pKa = 4.06WLFPTALEE485 pKa = 4.3WVDD488 pKa = 4.72GEE490 pKa = 4.47WKK492 pKa = 8.52VTNPVVEE499 pKa = 4.12IMVEE503 pKa = 3.97KK504 pKa = 9.62MASVDD509 pKa = 4.19IIFKK513 pKa = 9.99TPDD516 pKa = 3.05GNILEE521 pKa = 4.53DD522 pKa = 3.41WNYY525 pKa = 8.18TRR527 pKa = 11.84YY528 pKa = 10.06YY529 pKa = 10.24DD530 pKa = 4.04SEE532 pKa = 5.16GGGIEE537 pKa = 4.21TVPAPEE543 pKa = 4.89GYY545 pKa = 10.48EE546 pKa = 3.75FVGEE550 pKa = 3.7NTYY553 pKa = 10.93AVEE556 pKa = 4.48VIRR559 pKa = 11.84DD560 pKa = 3.63EE561 pKa = 4.28TGKK564 pKa = 10.75LVADD568 pKa = 4.18PSEE571 pKa = 3.92VTFWVKK577 pKa = 10.58EE578 pKa = 3.91IGSEE582 pKa = 4.29IPVTPSDD589 pKa = 3.49YY590 pKa = 11.03SFEE593 pKa = 4.1VKK595 pKa = 10.87YY596 pKa = 10.59IDD598 pKa = 4.83GEE600 pKa = 4.55GNPVPGGGTISFEE613 pKa = 3.83EE614 pKa = 4.06AGPYY618 pKa = 9.36NRR620 pKa = 11.84EE621 pKa = 4.74DD622 pKa = 3.13IPLPYY627 pKa = 10.21GYY629 pKa = 8.53MQIIPAHH636 pKa = 6.85PDD638 pKa = 3.31EE639 pKa = 5.76DD640 pKa = 3.98WLFPTSLVLVDD651 pKa = 4.98GEE653 pKa = 4.47WKK655 pKa = 8.46VTNPVVEE662 pKa = 4.12IMVEE666 pKa = 3.97KK667 pKa = 9.62MASVDD672 pKa = 5.05IIFQTPDD679 pKa = 2.95GNILEE684 pKa = 4.53DD685 pKa = 3.41WNYY688 pKa = 8.18TRR690 pKa = 11.84YY691 pKa = 10.06YY692 pKa = 10.24DD693 pKa = 4.04SEE695 pKa = 5.16GGGIEE700 pKa = 4.21TVPAPEE706 pKa = 4.77GYY708 pKa = 10.55EE709 pKa = 3.95FIGEE713 pKa = 3.68NTYY716 pKa = 10.89AVEE719 pKa = 4.48VIRR722 pKa = 11.84DD723 pKa = 3.63EE724 pKa = 4.28TGKK727 pKa = 10.75LVADD731 pKa = 4.06PSEE734 pKa = 4.12VIFIVKK740 pKa = 10.42AIDD743 pKa = 3.4TEE745 pKa = 4.88GPSDD749 pKa = 4.01PEE751 pKa = 4.22EE752 pKa = 4.38PSTPGGTNTPEE763 pKa = 4.34GNSPQTGDD771 pKa = 4.34DD772 pKa = 3.45SHH774 pKa = 8.32ASNLLLLILVSAGTLLLVTIIRR796 pKa = 11.84NQKK799 pKa = 8.33KK800 pKa = 9.75KK801 pKa = 11.06EE802 pKa = 3.95MTTEE806 pKa = 3.89EE807 pKa = 4.12
Molecular weight: 88.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0ED61|C0ED61_9FIRM UDP-galactose 4-epimerase OS=[Clostridium] methylpentosum DSM 5476 OX=537013 GN=CLOSTMETH_01787 PE=3 SV=1
MM1 pKa = 7.58LFFLRR6 pKa = 11.84FFISIHH12 pKa = 4.67TRR14 pKa = 11.84KK15 pKa = 8.61GTATRR20 pKa = 11.84RR21 pKa = 11.84GLSIAQTFLTISIHH35 pKa = 4.47TRR37 pKa = 11.84KK38 pKa = 8.8GTVTSRR44 pKa = 11.84SPGQIQRR51 pKa = 11.84AFSSIHH57 pKa = 4.49TRR59 pKa = 11.84KK60 pKa = 10.55GPGQLL65 pKa = 3.57
MM1 pKa = 7.58LFFLRR6 pKa = 11.84FFISIHH12 pKa = 4.67TRR14 pKa = 11.84KK15 pKa = 8.61GTATRR20 pKa = 11.84RR21 pKa = 11.84GLSIAQTFLTISIHH35 pKa = 4.47TRR37 pKa = 11.84KK38 pKa = 8.8GTVTSRR44 pKa = 11.84SPGQIQRR51 pKa = 11.84AFSSIHH57 pKa = 4.49TRR59 pKa = 11.84KK60 pKa = 10.55GPGQLL65 pKa = 3.57
Molecular weight: 7.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
988130 |
34 |
2451 |
252.9 |
28.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.249 ± 0.052 | 1.532 ± 0.022 |
5.58 ± 0.037 | 6.65 ± 0.043 |
4.258 ± 0.035 | 7.115 ± 0.039 |
1.732 ± 0.021 | 6.385 ± 0.041 |
5.662 ± 0.047 | 9.458 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.551 ± 0.018 | 4.234 ± 0.034 |
3.845 ± 0.028 | 4.066 ± 0.03 |
4.803 ± 0.044 | 6.535 ± 0.036 |
5.732 ± 0.047 | 6.794 ± 0.032 |
1.013 ± 0.018 | 3.803 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
