
Odobenus rosmarus divergens (Pacific walrus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Carnivora; Caniformia;
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
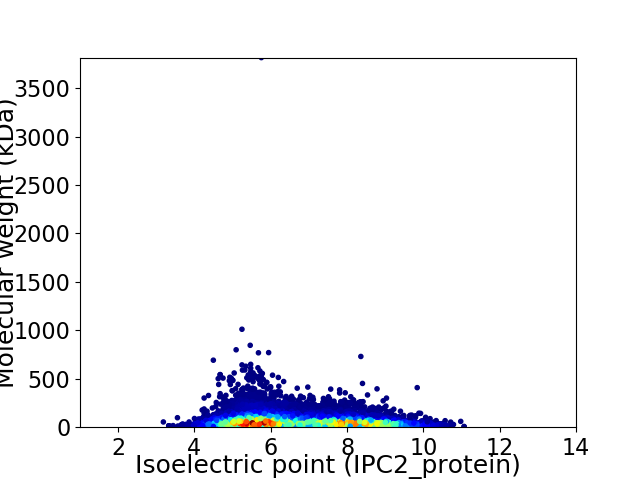
Virtual 2D-PAGE plot for 29573 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U3ZF50|A0A2U3ZF50_ODORO LOW QUALITY PROTEIN: uncharacterized protein C2orf91 homolog OS=Odobenus rosmarus divergens OX=9708 GN=LOC105757378 PE=4 SV=1
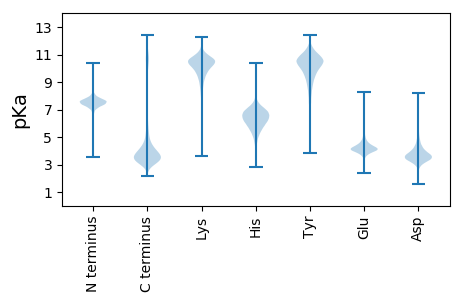
MM1 pKa = 7.22EE2 pKa = 5.11AVVLIPCVLGLLLLPLLAGALCVRR26 pKa = 11.84CRR28 pKa = 11.84EE29 pKa = 4.25LPGSYY34 pKa = 8.64DD35 pKa = 3.36TTASDD40 pKa = 3.85SLTPSSIVLKK50 pKa = 10.42RR51 pKa = 11.84PPTVVPWPSATSYY64 pKa = 11.6APVTSYY70 pKa = 11.28PSLSQPDD77 pKa = 4.54LLPIPRR83 pKa = 11.84SPQPHH88 pKa = 6.34HH89 pKa = 6.86VPSSRR94 pKa = 11.84QDD96 pKa = 3.32SEE98 pKa = 4.34GANSVASYY106 pKa = 10.76EE107 pKa = 4.13NEE109 pKa = 3.86EE110 pKa = 4.21PVCEE114 pKa = 4.29DD115 pKa = 3.85EE116 pKa = 4.82EE117 pKa = 4.97EE118 pKa = 4.14EE119 pKa = 5.45DD120 pKa = 3.52EE121 pKa = 5.4DD122 pKa = 4.19YY123 pKa = 11.5HH124 pKa = 8.35NEE126 pKa = 4.13GYY128 pKa = 11.01LEE130 pKa = 4.22VLPDD134 pKa = 3.6STPAPSSAVPPAPVLSNPGLRR155 pKa = 11.84DD156 pKa = 3.09SAFSMEE162 pKa = 4.23SGEE165 pKa = 4.92DD166 pKa = 3.56YY167 pKa = 11.72VNVLEE172 pKa = 4.69SEE174 pKa = 4.33EE175 pKa = 4.25SADD178 pKa = 3.65VSLDD182 pKa = 3.29GSRR185 pKa = 11.84EE186 pKa = 3.96YY187 pKa = 11.72VNVSQEE193 pKa = 4.41LPPMAKK199 pKa = 8.92TEE201 pKa = 3.92PAILSSQKK209 pKa = 10.17VEE211 pKa = 4.63DD212 pKa = 4.1EE213 pKa = 4.25DD214 pKa = 5.59DD215 pKa = 3.99EE216 pKa = 4.77EE217 pKa = 5.11EE218 pKa = 4.32EE219 pKa = 4.72GAPDD223 pKa = 4.17YY224 pKa = 11.56EE225 pKa = 4.37NLL227 pKa = 3.83
MM1 pKa = 7.22EE2 pKa = 5.11AVVLIPCVLGLLLLPLLAGALCVRR26 pKa = 11.84CRR28 pKa = 11.84EE29 pKa = 4.25LPGSYY34 pKa = 8.64DD35 pKa = 3.36TTASDD40 pKa = 3.85SLTPSSIVLKK50 pKa = 10.42RR51 pKa = 11.84PPTVVPWPSATSYY64 pKa = 11.6APVTSYY70 pKa = 11.28PSLSQPDD77 pKa = 4.54LLPIPRR83 pKa = 11.84SPQPHH88 pKa = 6.34HH89 pKa = 6.86VPSSRR94 pKa = 11.84QDD96 pKa = 3.32SEE98 pKa = 4.34GANSVASYY106 pKa = 10.76EE107 pKa = 4.13NEE109 pKa = 3.86EE110 pKa = 4.21PVCEE114 pKa = 4.29DD115 pKa = 3.85EE116 pKa = 4.82EE117 pKa = 4.97EE118 pKa = 4.14EE119 pKa = 5.45DD120 pKa = 3.52EE121 pKa = 5.4DD122 pKa = 4.19YY123 pKa = 11.5HH124 pKa = 8.35NEE126 pKa = 4.13GYY128 pKa = 11.01LEE130 pKa = 4.22VLPDD134 pKa = 3.6STPAPSSAVPPAPVLSNPGLRR155 pKa = 11.84DD156 pKa = 3.09SAFSMEE162 pKa = 4.23SGEE165 pKa = 4.92DD166 pKa = 3.56YY167 pKa = 11.72VNVLEE172 pKa = 4.69SEE174 pKa = 4.33EE175 pKa = 4.25SADD178 pKa = 3.65VSLDD182 pKa = 3.29GSRR185 pKa = 11.84EE186 pKa = 3.96YY187 pKa = 11.72VNVSQEE193 pKa = 4.41LPPMAKK199 pKa = 8.92TEE201 pKa = 3.92PAILSSQKK209 pKa = 10.17VEE211 pKa = 4.63DD212 pKa = 4.1EE213 pKa = 4.25DD214 pKa = 5.59DD215 pKa = 3.99EE216 pKa = 4.77EE217 pKa = 5.11EE218 pKa = 4.32EE219 pKa = 4.72GAPDD223 pKa = 4.17YY224 pKa = 11.56EE225 pKa = 4.37NLL227 pKa = 3.83
Molecular weight: 24.48 kDa
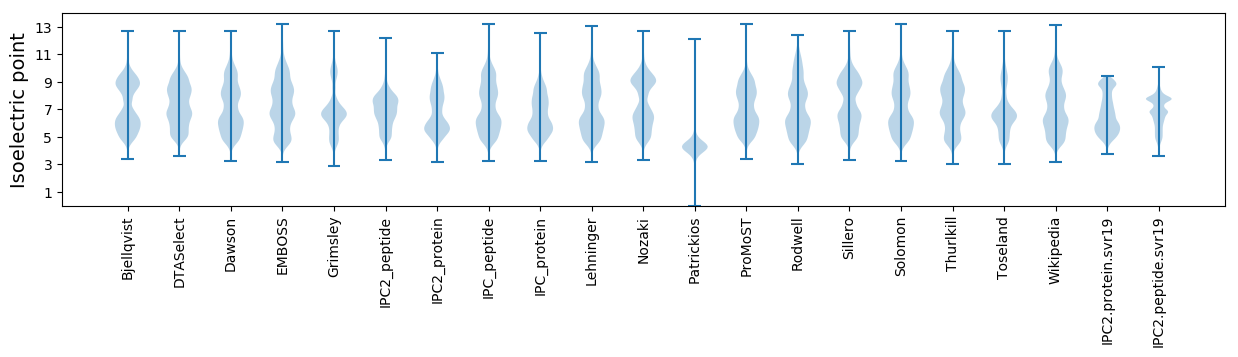
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U3VB18|A0A2U3VB18_ODORO mycophenolic acid acyl-glucuronide esterase mitochondrial OS=Odobenus rosmarus divergens OX=9708 GN=ABHD10 PE=4 SV=1
MM1 pKa = 7.35QSVCVPPLPHH11 pKa = 6.93LLVGSFYY18 pKa = 10.24PFPVLPTAQCPVPLPPSSPGSQPSGRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84SRR48 pKa = 11.84ASPSGHH54 pKa = 6.81ASQPGPALRR63 pKa = 11.84SQARR67 pKa = 11.84ASGPVLRR74 pKa = 11.84SQARR78 pKa = 11.84ALGPVLRR85 pKa = 11.84SQAARR90 pKa = 11.84PGPALNGRR98 pKa = 11.84ATPPGTAFRR107 pKa = 11.84GHH109 pKa = 6.72ASGLGSALCSRR120 pKa = 11.84ASAPGPARR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84GAPAPGPALRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84HH143 pKa = 5.49VPVSGPALRR152 pKa = 11.84SRR154 pKa = 11.84SRR156 pKa = 11.84IRR158 pKa = 11.84GRR160 pKa = 11.84TSSSGAARR168 pKa = 11.84RR169 pKa = 11.84NRR171 pKa = 11.84GRR173 pKa = 11.84APAPGPALRR182 pKa = 11.84SRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84SSSPGAARR194 pKa = 11.84RR195 pKa = 11.84SRR197 pKa = 11.84GRR199 pKa = 11.84APAPGPALRR208 pKa = 11.84TLRR211 pKa = 11.84SRR213 pKa = 11.84RR214 pKa = 11.84RR215 pKa = 11.84SSSPGAARR223 pKa = 11.84RR224 pKa = 11.84SRR226 pKa = 11.84GRR228 pKa = 11.84APAPGPALRR237 pKa = 11.84SRR239 pKa = 11.84RR240 pKa = 11.84RR241 pKa = 11.84SSSPGAARR249 pKa = 11.84RR250 pKa = 11.84SRR252 pKa = 11.84GRR254 pKa = 11.84APAPGPALRR263 pKa = 11.84SRR265 pKa = 11.84HH266 pKa = 5.43SSSPGAASRR275 pKa = 11.84SRR277 pKa = 11.84GRR279 pKa = 11.84APAPGAASRR288 pKa = 11.84SRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84APASGPARR300 pKa = 11.84RR301 pKa = 11.84SRR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84SSSPGATRR313 pKa = 11.84RR314 pKa = 11.84SRR316 pKa = 11.84GRR318 pKa = 11.84ASAPGPALRR327 pKa = 11.84SRR329 pKa = 11.84RR330 pKa = 11.84RR331 pKa = 11.84SSSPGSARR339 pKa = 11.84RR340 pKa = 11.84SRR342 pKa = 11.84GRR344 pKa = 11.84VSARR348 pKa = 11.84GPALRR353 pKa = 11.84SRR355 pKa = 11.84RR356 pKa = 11.84SSSPGPARR364 pKa = 11.84RR365 pKa = 11.84SRR367 pKa = 11.84ASAPGPAYY375 pKa = 10.51RR376 pKa = 11.84NRR378 pKa = 11.84ASSSRR383 pKa = 11.84ASGPSSALNNQASPPGPALRR403 pKa = 11.84NRR405 pKa = 11.84ASGSGPALRR414 pKa = 11.84NGCPTPGFVLRR425 pKa = 11.84GRR427 pKa = 11.84TIQRR431 pKa = 11.84SFPRR435 pKa = 11.84SRR437 pKa = 11.84ASLPVPPGPSPPSSPGVGLRR457 pKa = 11.84RR458 pKa = 11.84LVWQSSSSSPNPEE471 pKa = 3.95VPSLGAQPRR480 pKa = 11.84WHH482 pKa = 6.76AVRR485 pKa = 11.84MRR487 pKa = 11.84ASSPSPPGRR496 pKa = 11.84LLPFTEE502 pKa = 5.45LYY504 pKa = 10.83GEE506 pKa = 4.47SSSSSSSTSSSGSPGQSPSSSSSSTSSPKK535 pKa = 10.06FCGLGSISTPSPASLRR551 pKa = 11.84RR552 pKa = 11.84ALLPEE557 pKa = 4.99LDD559 pKa = 4.03ALSPLSPGEE568 pKa = 3.88EE569 pKa = 4.44DD570 pKa = 3.44EE571 pKa = 4.5TGSMPSSPTPPVVV584 pKa = 2.94
MM1 pKa = 7.35QSVCVPPLPHH11 pKa = 6.93LLVGSFYY18 pKa = 10.24PFPVLPTAQCPVPLPPSSPGSQPSGRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84SRR48 pKa = 11.84ASPSGHH54 pKa = 6.81ASQPGPALRR63 pKa = 11.84SQARR67 pKa = 11.84ASGPVLRR74 pKa = 11.84SQARR78 pKa = 11.84ALGPVLRR85 pKa = 11.84SQAARR90 pKa = 11.84PGPALNGRR98 pKa = 11.84ATPPGTAFRR107 pKa = 11.84GHH109 pKa = 6.72ASGLGSALCSRR120 pKa = 11.84ASAPGPARR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84GAPAPGPALRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84HH143 pKa = 5.49VPVSGPALRR152 pKa = 11.84SRR154 pKa = 11.84SRR156 pKa = 11.84IRR158 pKa = 11.84GRR160 pKa = 11.84TSSSGAARR168 pKa = 11.84RR169 pKa = 11.84NRR171 pKa = 11.84GRR173 pKa = 11.84APAPGPALRR182 pKa = 11.84SRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84SSSPGAARR194 pKa = 11.84RR195 pKa = 11.84SRR197 pKa = 11.84GRR199 pKa = 11.84APAPGPALRR208 pKa = 11.84TLRR211 pKa = 11.84SRR213 pKa = 11.84RR214 pKa = 11.84RR215 pKa = 11.84SSSPGAARR223 pKa = 11.84RR224 pKa = 11.84SRR226 pKa = 11.84GRR228 pKa = 11.84APAPGPALRR237 pKa = 11.84SRR239 pKa = 11.84RR240 pKa = 11.84RR241 pKa = 11.84SSSPGAARR249 pKa = 11.84RR250 pKa = 11.84SRR252 pKa = 11.84GRR254 pKa = 11.84APAPGPALRR263 pKa = 11.84SRR265 pKa = 11.84HH266 pKa = 5.43SSSPGAASRR275 pKa = 11.84SRR277 pKa = 11.84GRR279 pKa = 11.84APAPGAASRR288 pKa = 11.84SRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84APASGPARR300 pKa = 11.84RR301 pKa = 11.84SRR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84SSSPGATRR313 pKa = 11.84RR314 pKa = 11.84SRR316 pKa = 11.84GRR318 pKa = 11.84ASAPGPALRR327 pKa = 11.84SRR329 pKa = 11.84RR330 pKa = 11.84RR331 pKa = 11.84SSSPGSARR339 pKa = 11.84RR340 pKa = 11.84SRR342 pKa = 11.84GRR344 pKa = 11.84VSARR348 pKa = 11.84GPALRR353 pKa = 11.84SRR355 pKa = 11.84RR356 pKa = 11.84SSSPGPARR364 pKa = 11.84RR365 pKa = 11.84SRR367 pKa = 11.84ASAPGPAYY375 pKa = 10.51RR376 pKa = 11.84NRR378 pKa = 11.84ASSSRR383 pKa = 11.84ASGPSSALNNQASPPGPALRR403 pKa = 11.84NRR405 pKa = 11.84ASGSGPALRR414 pKa = 11.84NGCPTPGFVLRR425 pKa = 11.84GRR427 pKa = 11.84TIQRR431 pKa = 11.84SFPRR435 pKa = 11.84SRR437 pKa = 11.84ASLPVPPGPSPPSSPGVGLRR457 pKa = 11.84RR458 pKa = 11.84LVWQSSSSSPNPEE471 pKa = 3.95VPSLGAQPRR480 pKa = 11.84WHH482 pKa = 6.76AVRR485 pKa = 11.84MRR487 pKa = 11.84ASSPSPPGRR496 pKa = 11.84LLPFTEE502 pKa = 5.45LYY504 pKa = 10.83GEE506 pKa = 4.47SSSSSSSTSSSGSPGQSPSSSSSSTSSPKK535 pKa = 10.06FCGLGSISTPSPASLRR551 pKa = 11.84RR552 pKa = 11.84ALLPEE557 pKa = 4.99LDD559 pKa = 4.03ALSPLSPGEE568 pKa = 3.88EE569 pKa = 4.44DD570 pKa = 3.44EE571 pKa = 4.5TGSMPSSPTPPVVV584 pKa = 2.94
Molecular weight: 59.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17273837 |
31 |
34340 |
584.1 |
65.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.073 ± 0.015 | 2.162 ± 0.013 |
4.864 ± 0.009 | 7.116 ± 0.018 |
3.64 ± 0.01 | 6.624 ± 0.019 |
2.568 ± 0.007 | 4.369 ± 0.012 |
5.724 ± 0.018 | 9.843 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.184 ± 0.007 | 3.622 ± 0.009 |
6.337 ± 0.021 | 4.734 ± 0.013 |
5.738 ± 0.014 | 8.263 ± 0.017 |
5.25 ± 0.013 | 6.022 ± 0.012 |
1.177 ± 0.005 | 2.676 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
