
Halomonas sp. PA5
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; unclassified Halomonas
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
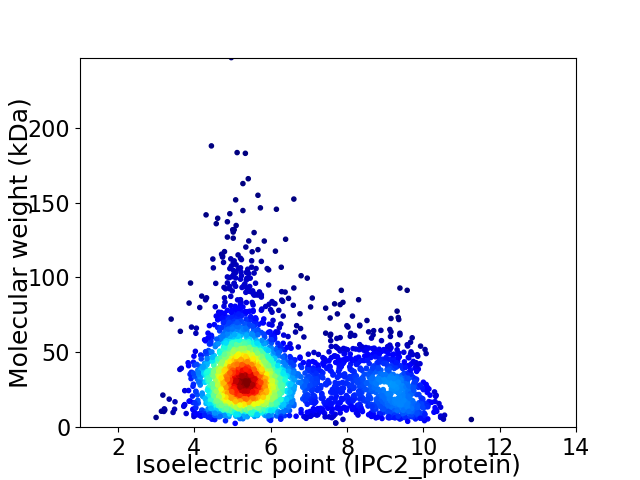
Virtual 2D-PAGE plot for 3407 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M4FNX0|A0A6M4FNX0_9GAMM Acylphosphatase OS=Halomonas sp. PA5 OX=2730357 GN=HIO72_16250 PE=3 SV=1
MM1 pKa = 7.7PRR3 pKa = 11.84TLPNRR8 pKa = 11.84SILAATLSTTLLATALSAPMTQAQEE33 pKa = 3.72RR34 pKa = 11.84DD35 pKa = 3.59QFSVCWSIYY44 pKa = 9.54AGWMPWAYY52 pKa = 10.68ADD54 pKa = 4.53IEE56 pKa = 4.67GIVDD60 pKa = 3.57KK61 pKa = 10.6WADD64 pKa = 3.59EE65 pKa = 4.13YY66 pKa = 11.54GISIDD71 pKa = 3.78VIQVNDD77 pKa = 3.61YY78 pKa = 11.01VEE80 pKa = 5.11SINQFTSGNVDD91 pKa = 3.21GCAMTNMDD99 pKa = 5.17ALTIPAASGVDD110 pKa = 3.58STALIINDD118 pKa = 3.79YY119 pKa = 11.35SDD121 pKa = 4.12GNDD124 pKa = 4.04GIVLKK129 pKa = 11.11NSDD132 pKa = 4.49DD133 pKa = 3.81LADD136 pKa = 3.49IEE138 pKa = 4.33GRR140 pKa = 11.84RR141 pKa = 11.84VNLVQFSVSHH151 pKa = 5.55YY152 pKa = 8.95FLARR156 pKa = 11.84ALEE159 pKa = 4.48SVGLTEE165 pKa = 5.89RR166 pKa = 11.84DD167 pKa = 3.11IEE169 pKa = 4.52TVNTSDD175 pKa = 3.85ADD177 pKa = 3.6IVGLFSSASTEE188 pKa = 5.17AVAAWNPQLSAIADD202 pKa = 3.8MPDD205 pKa = 3.29SNVVYY210 pKa = 9.59TSAEE214 pKa = 3.85IPGEE218 pKa = 4.05ILDD221 pKa = 3.86MMVVNTDD228 pKa = 3.31TLAANPDD235 pKa = 3.58FGHH238 pKa = 6.83ALVGAWYY245 pKa = 9.75EE246 pKa = 4.12VMALVEE252 pKa = 4.49ARR254 pKa = 11.84DD255 pKa = 3.84EE256 pKa = 4.13QALSIMADD264 pKa = 3.14AAGTDD269 pKa = 3.62LEE271 pKa = 5.34GYY273 pKa = 9.63LAQLDD278 pKa = 3.86ATYY281 pKa = 10.47LYY283 pKa = 10.01TDD285 pKa = 3.55PTEE288 pKa = 5.49AIALMEE294 pKa = 4.51GDD296 pKa = 4.18EE297 pKa = 4.87LLEE300 pKa = 4.04TMQRR304 pKa = 11.84VAEE307 pKa = 4.28FSFEE311 pKa = 4.11HH312 pKa = 6.7GLLGDD317 pKa = 4.01MAPDD321 pKa = 4.04PGVVGVEE328 pKa = 4.5TPSGVYY334 pKa = 10.35GDD336 pKa = 4.77EE337 pKa = 4.3SNIMLRR343 pKa = 11.84FDD345 pKa = 3.72PSYY348 pKa = 8.2TQAYY352 pKa = 8.53IDD354 pKa = 3.71SRR356 pKa = 11.84QQ357 pKa = 3.03
MM1 pKa = 7.7PRR3 pKa = 11.84TLPNRR8 pKa = 11.84SILAATLSTTLLATALSAPMTQAQEE33 pKa = 3.72RR34 pKa = 11.84DD35 pKa = 3.59QFSVCWSIYY44 pKa = 9.54AGWMPWAYY52 pKa = 10.68ADD54 pKa = 4.53IEE56 pKa = 4.67GIVDD60 pKa = 3.57KK61 pKa = 10.6WADD64 pKa = 3.59EE65 pKa = 4.13YY66 pKa = 11.54GISIDD71 pKa = 3.78VIQVNDD77 pKa = 3.61YY78 pKa = 11.01VEE80 pKa = 5.11SINQFTSGNVDD91 pKa = 3.21GCAMTNMDD99 pKa = 5.17ALTIPAASGVDD110 pKa = 3.58STALIINDD118 pKa = 3.79YY119 pKa = 11.35SDD121 pKa = 4.12GNDD124 pKa = 4.04GIVLKK129 pKa = 11.11NSDD132 pKa = 4.49DD133 pKa = 3.81LADD136 pKa = 3.49IEE138 pKa = 4.33GRR140 pKa = 11.84RR141 pKa = 11.84VNLVQFSVSHH151 pKa = 5.55YY152 pKa = 8.95FLARR156 pKa = 11.84ALEE159 pKa = 4.48SVGLTEE165 pKa = 5.89RR166 pKa = 11.84DD167 pKa = 3.11IEE169 pKa = 4.52TVNTSDD175 pKa = 3.85ADD177 pKa = 3.6IVGLFSSASTEE188 pKa = 5.17AVAAWNPQLSAIADD202 pKa = 3.8MPDD205 pKa = 3.29SNVVYY210 pKa = 9.59TSAEE214 pKa = 3.85IPGEE218 pKa = 4.05ILDD221 pKa = 3.86MMVVNTDD228 pKa = 3.31TLAANPDD235 pKa = 3.58FGHH238 pKa = 6.83ALVGAWYY245 pKa = 9.75EE246 pKa = 4.12VMALVEE252 pKa = 4.49ARR254 pKa = 11.84DD255 pKa = 3.84EE256 pKa = 4.13QALSIMADD264 pKa = 3.14AAGTDD269 pKa = 3.62LEE271 pKa = 5.34GYY273 pKa = 9.63LAQLDD278 pKa = 3.86ATYY281 pKa = 10.47LYY283 pKa = 10.01TDD285 pKa = 3.55PTEE288 pKa = 5.49AIALMEE294 pKa = 4.51GDD296 pKa = 4.18EE297 pKa = 4.87LLEE300 pKa = 4.04TMQRR304 pKa = 11.84VAEE307 pKa = 4.28FSFEE311 pKa = 4.11HH312 pKa = 6.7GLLGDD317 pKa = 4.01MAPDD321 pKa = 4.04PGVVGVEE328 pKa = 4.5TPSGVYY334 pKa = 10.35GDD336 pKa = 4.77EE337 pKa = 4.3SNIMLRR343 pKa = 11.84FDD345 pKa = 3.72PSYY348 pKa = 8.2TQAYY352 pKa = 8.53IDD354 pKa = 3.71SRR356 pKa = 11.84QQ357 pKa = 3.03
Molecular weight: 38.57 kDa
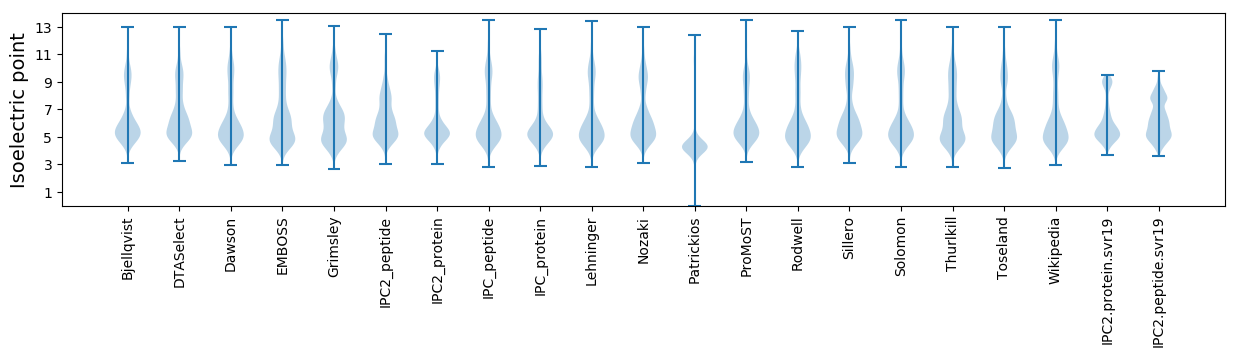
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M4FKT5|A0A6M4FKT5_9GAMM Exodeoxyribonuclease 7 large subunit OS=Halomonas sp. PA5 OX=2730357 GN=xseA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1107370 |
23 |
2263 |
325.0 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.004 ± 0.046 | 0.955 ± 0.013 |
5.446 ± 0.041 | 6.68 ± 0.047 |
3.501 ± 0.026 | 8.158 ± 0.043 |
2.469 ± 0.022 | 4.987 ± 0.03 |
2.639 ± 0.032 | 11.556 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.631 ± 0.02 | 2.58 ± 0.022 |
4.873 ± 0.025 | 3.89 ± 0.032 |
7.272 ± 0.043 | 5.635 ± 0.028 |
4.868 ± 0.025 | 7.027 ± 0.037 |
1.442 ± 0.019 | 2.388 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
