
Leptonychotes weddellii (Weddell seal) (Otaria weddellii)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Carnivora;
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
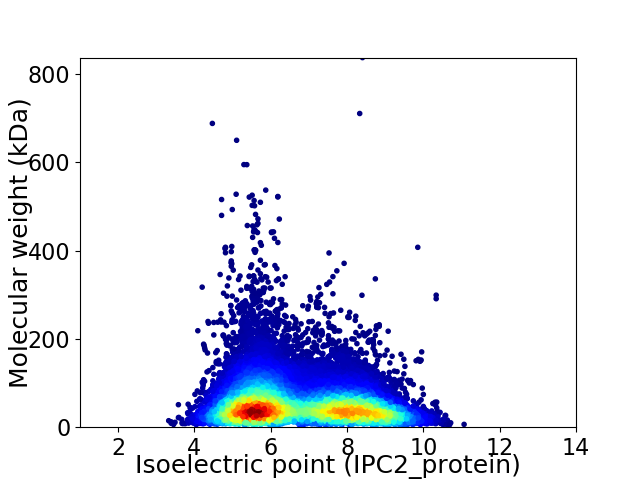
Virtual 2D-PAGE plot for 34224 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U3YBR7|A0A2U3YBR7_LEPWE PAK4-inhibitor INKA2 OS=Leptonychotes weddellii OX=9713 GN=INKA2 PE=3 SV=1
LL1 pKa = 8.17VCILCSQDD9 pKa = 3.35NCPLLPNSGQEE20 pKa = 4.8DD21 pKa = 4.05FDD23 pKa = 5.49KK24 pKa = 11.33DD25 pKa = 3.82GTGDD29 pKa = 3.68ACDD32 pKa = 4.45EE33 pKa = 4.97DD34 pKa = 5.18DD35 pKa = 6.3DD36 pKa = 5.46NDD38 pKa = 4.45GVDD41 pKa = 4.91DD42 pKa = 5.27EE43 pKa = 5.57KK44 pKa = 11.77DD45 pKa = 3.31NCQLLFNPRR54 pKa = 11.84QFDD57 pKa = 3.57YY58 pKa = 11.45DD59 pKa = 3.52KK60 pKa = 11.69DD61 pKa = 4.14EE62 pKa = 4.7VGDD65 pKa = 3.79RR66 pKa = 11.84CDD68 pKa = 3.2NCPYY72 pKa = 10.11VHH74 pKa = 6.82NPAQIDD80 pKa = 3.5TDD82 pKa = 3.98NNGEE86 pKa = 3.91GDD88 pKa = 3.81ACSVDD93 pKa = 3.02IDD95 pKa = 4.08GDD97 pKa = 4.08DD98 pKa = 3.82VFNEE102 pKa = 4.26RR103 pKa = 11.84DD104 pKa = 3.43NCPYY108 pKa = 10.72VYY110 pKa = 9.89NTDD113 pKa = 3.55QRR115 pKa = 11.84DD116 pKa = 3.41TDD118 pKa = 3.67GDD120 pKa = 4.33GVGDD124 pKa = 3.92HH125 pKa = 7.43CDD127 pKa = 3.4NCPLVHH133 pKa = 6.91NPDD136 pKa = 3.37QTDD139 pKa = 2.77VDD141 pKa = 3.78NDD143 pKa = 4.03LVGDD147 pKa = 3.72QCDD150 pKa = 3.64NNEE153 pKa = 5.1DD154 pKa = 3.49IDD156 pKa = 4.85EE157 pKa = 5.38DD158 pKa = 3.95GHH160 pKa = 6.99QNNQDD165 pKa = 3.17NCPYY169 pKa = 10.33ISNANQADD177 pKa = 3.98HH178 pKa = 7.28DD179 pKa = 4.49HH180 pKa = 7.58DD181 pKa = 4.53GQGDD185 pKa = 3.81ACDD188 pKa = 4.6SDD190 pKa = 4.48DD191 pKa = 6.02DD192 pKa = 4.76NDD194 pKa = 4.26GVPDD198 pKa = 4.51DD199 pKa = 4.98RR200 pKa = 11.84DD201 pKa = 3.34NCRR204 pKa = 11.84LVSNPGQEE212 pKa = 5.02DD213 pKa = 3.49SDD215 pKa = 4.2GDD217 pKa = 4.03GRR219 pKa = 11.84GDD221 pKa = 3.29ACKK224 pKa = 10.67DD225 pKa = 3.34DD226 pKa = 4.2FDD228 pKa = 4.98NDD230 pKa = 4.83RR231 pKa = 11.84IPDD234 pKa = 3.63IDD236 pKa = 4.49DD237 pKa = 3.64VCPEE241 pKa = 3.75NSAISEE247 pKa = 4.02TDD249 pKa = 3.4FRR251 pKa = 11.84NFQMVHH257 pKa = 7.1LDD259 pKa = 3.63PKK261 pKa = 10.23GTTQIDD267 pKa = 4.0PNWVIRR273 pKa = 11.84HH274 pKa = 4.99QGKK277 pKa = 9.11EE278 pKa = 3.87LVQTANSDD286 pKa = 3.28PGIAVGFDD294 pKa = 3.2EE295 pKa = 5.37FGSVDD300 pKa = 3.7FSGTFYY306 pKa = 11.53VNTDD310 pKa = 3.3RR311 pKa = 11.84DD312 pKa = 3.36DD313 pKa = 4.91DD314 pKa = 4.04YY315 pKa = 12.01AGFVFGYY322 pKa = 9.43QSSSRR327 pKa = 11.84FYY329 pKa = 10.25VVMWKK334 pKa = 9.82QVTQTYY340 pKa = 8.78WEE342 pKa = 4.39DD343 pKa = 3.58QPTRR347 pKa = 11.84AYY349 pKa = 10.06GYY351 pKa = 9.91SGVSLKK357 pKa = 10.51VVNSTTGTGEE367 pKa = 4.03HH368 pKa = 6.41LRR370 pKa = 11.84NALWHH375 pKa = 5.99TGNTEE380 pKa = 4.02GQVRR384 pKa = 11.84TLWHH388 pKa = 6.69DD389 pKa = 3.57PKK391 pKa = 11.28NIGWKK396 pKa = 10.31DD397 pKa = 3.26YY398 pKa = 9.58TAYY401 pKa = 10.32RR402 pKa = 11.84WHH404 pKa = 6.5LTHH407 pKa = 7.17RR408 pKa = 11.84PKK410 pKa = 10.37TGYY413 pKa = 9.82IRR415 pKa = 11.84VLVHH419 pKa = 6.46EE420 pKa = 5.84GKK422 pKa = 9.99QVMADD427 pKa = 3.41SGPVYY432 pKa = 10.46DD433 pKa = 3.47QTYY436 pKa = 9.92AGGRR440 pKa = 11.84LGLFVFSQEE449 pKa = 3.65MVYY452 pKa = 10.62FSDD455 pKa = 4.84LKK457 pKa = 10.82YY458 pKa = 10.49EE459 pKa = 4.28CRR461 pKa = 11.84DD462 pKa = 3.3VV463 pKa = 3.44
LL1 pKa = 8.17VCILCSQDD9 pKa = 3.35NCPLLPNSGQEE20 pKa = 4.8DD21 pKa = 4.05FDD23 pKa = 5.49KK24 pKa = 11.33DD25 pKa = 3.82GTGDD29 pKa = 3.68ACDD32 pKa = 4.45EE33 pKa = 4.97DD34 pKa = 5.18DD35 pKa = 6.3DD36 pKa = 5.46NDD38 pKa = 4.45GVDD41 pKa = 4.91DD42 pKa = 5.27EE43 pKa = 5.57KK44 pKa = 11.77DD45 pKa = 3.31NCQLLFNPRR54 pKa = 11.84QFDD57 pKa = 3.57YY58 pKa = 11.45DD59 pKa = 3.52KK60 pKa = 11.69DD61 pKa = 4.14EE62 pKa = 4.7VGDD65 pKa = 3.79RR66 pKa = 11.84CDD68 pKa = 3.2NCPYY72 pKa = 10.11VHH74 pKa = 6.82NPAQIDD80 pKa = 3.5TDD82 pKa = 3.98NNGEE86 pKa = 3.91GDD88 pKa = 3.81ACSVDD93 pKa = 3.02IDD95 pKa = 4.08GDD97 pKa = 4.08DD98 pKa = 3.82VFNEE102 pKa = 4.26RR103 pKa = 11.84DD104 pKa = 3.43NCPYY108 pKa = 10.72VYY110 pKa = 9.89NTDD113 pKa = 3.55QRR115 pKa = 11.84DD116 pKa = 3.41TDD118 pKa = 3.67GDD120 pKa = 4.33GVGDD124 pKa = 3.92HH125 pKa = 7.43CDD127 pKa = 3.4NCPLVHH133 pKa = 6.91NPDD136 pKa = 3.37QTDD139 pKa = 2.77VDD141 pKa = 3.78NDD143 pKa = 4.03LVGDD147 pKa = 3.72QCDD150 pKa = 3.64NNEE153 pKa = 5.1DD154 pKa = 3.49IDD156 pKa = 4.85EE157 pKa = 5.38DD158 pKa = 3.95GHH160 pKa = 6.99QNNQDD165 pKa = 3.17NCPYY169 pKa = 10.33ISNANQADD177 pKa = 3.98HH178 pKa = 7.28DD179 pKa = 4.49HH180 pKa = 7.58DD181 pKa = 4.53GQGDD185 pKa = 3.81ACDD188 pKa = 4.6SDD190 pKa = 4.48DD191 pKa = 6.02DD192 pKa = 4.76NDD194 pKa = 4.26GVPDD198 pKa = 4.51DD199 pKa = 4.98RR200 pKa = 11.84DD201 pKa = 3.34NCRR204 pKa = 11.84LVSNPGQEE212 pKa = 5.02DD213 pKa = 3.49SDD215 pKa = 4.2GDD217 pKa = 4.03GRR219 pKa = 11.84GDD221 pKa = 3.29ACKK224 pKa = 10.67DD225 pKa = 3.34DD226 pKa = 4.2FDD228 pKa = 4.98NDD230 pKa = 4.83RR231 pKa = 11.84IPDD234 pKa = 3.63IDD236 pKa = 4.49DD237 pKa = 3.64VCPEE241 pKa = 3.75NSAISEE247 pKa = 4.02TDD249 pKa = 3.4FRR251 pKa = 11.84NFQMVHH257 pKa = 7.1LDD259 pKa = 3.63PKK261 pKa = 10.23GTTQIDD267 pKa = 4.0PNWVIRR273 pKa = 11.84HH274 pKa = 4.99QGKK277 pKa = 9.11EE278 pKa = 3.87LVQTANSDD286 pKa = 3.28PGIAVGFDD294 pKa = 3.2EE295 pKa = 5.37FGSVDD300 pKa = 3.7FSGTFYY306 pKa = 11.53VNTDD310 pKa = 3.3RR311 pKa = 11.84DD312 pKa = 3.36DD313 pKa = 4.91DD314 pKa = 4.04YY315 pKa = 12.01AGFVFGYY322 pKa = 9.43QSSSRR327 pKa = 11.84FYY329 pKa = 10.25VVMWKK334 pKa = 9.82QVTQTYY340 pKa = 8.78WEE342 pKa = 4.39DD343 pKa = 3.58QPTRR347 pKa = 11.84AYY349 pKa = 10.06GYY351 pKa = 9.91SGVSLKK357 pKa = 10.51VVNSTTGTGEE367 pKa = 4.03HH368 pKa = 6.41LRR370 pKa = 11.84NALWHH375 pKa = 5.99TGNTEE380 pKa = 4.02GQVRR384 pKa = 11.84TLWHH388 pKa = 6.69DD389 pKa = 3.57PKK391 pKa = 11.28NIGWKK396 pKa = 10.31DD397 pKa = 3.26YY398 pKa = 9.58TAYY401 pKa = 10.32RR402 pKa = 11.84WHH404 pKa = 6.5LTHH407 pKa = 7.17RR408 pKa = 11.84PKK410 pKa = 10.37TGYY413 pKa = 9.82IRR415 pKa = 11.84VLVHH419 pKa = 6.46EE420 pKa = 5.84GKK422 pKa = 9.99QVMADD427 pKa = 3.41SGPVYY432 pKa = 10.46DD433 pKa = 3.47QTYY436 pKa = 9.92AGGRR440 pKa = 11.84LGLFVFSQEE449 pKa = 3.65MVYY452 pKa = 10.62FSDD455 pKa = 4.84LKK457 pKa = 10.82YY458 pKa = 10.49EE459 pKa = 4.28CRR461 pKa = 11.84DD462 pKa = 3.3VV463 pKa = 3.44
Molecular weight: 51.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7F8QEU1|A0A7F8QEU1_LEPWE sodium/hydrogen exchanger 8 OS=Leptonychotes weddellii OX=9713 GN=LOC115938833 PE=3 SV=1
RR1 pKa = 7.42PRR3 pKa = 11.84NAQSLPQKK11 pKa = 9.78PRR13 pKa = 11.84NAQSLPQRR21 pKa = 11.84PQNAQNLPQRR31 pKa = 11.84PQNIVRR37 pKa = 11.84LPRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84NAQSLPRR50 pKa = 11.84RR51 pKa = 11.84AQNILSLPQRR61 pKa = 11.84PRR63 pKa = 11.84NAQSLPQRR71 pKa = 11.84PQNTQNLPQRR81 pKa = 11.84PRR83 pKa = 11.84NIVNLPRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84NIVSLPGRR101 pKa = 11.84LRR103 pKa = 11.84NAQSLPWRR111 pKa = 11.84PRR113 pKa = 11.84NILNLPQRR121 pKa = 11.84PRR123 pKa = 11.84NAQSLPRR130 pKa = 11.84KK131 pKa = 8.74PRR133 pKa = 11.84NAQSLSRR140 pKa = 11.84RR141 pKa = 11.84PLNTQSLPRR150 pKa = 11.84GPRR153 pKa = 11.84NAQSLPQRR161 pKa = 11.84PWNAQSLPCGSQNAQSLSQRR181 pKa = 11.84PWNAQSLPQGSRR193 pKa = 11.84SILSLPRR200 pKa = 11.84KK201 pKa = 9.26LRR203 pKa = 11.84NAQSLPRR210 pKa = 11.84RR211 pKa = 11.84PQNTQSLQQKK221 pKa = 7.64PQNILSLSQRR231 pKa = 11.84PQNAQNLPRR240 pKa = 11.84RR241 pKa = 11.84PQNILSLPQKK251 pKa = 9.74PRR253 pKa = 11.84NAQSLPRR260 pKa = 11.84RR261 pKa = 11.84PQNTQSLPRR270 pKa = 11.84RR271 pKa = 11.84PQNAQNLPQRR281 pKa = 11.84PQNAQSLPQRR291 pKa = 11.84PQNTQNLQRR300 pKa = 11.84RR301 pKa = 11.84PQNAQNLPQGPQNILSLPQRR321 pKa = 11.84PQNAQSLPP329 pKa = 3.6
RR1 pKa = 7.42PRR3 pKa = 11.84NAQSLPQKK11 pKa = 9.78PRR13 pKa = 11.84NAQSLPQRR21 pKa = 11.84PQNAQNLPQRR31 pKa = 11.84PQNIVRR37 pKa = 11.84LPRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84NAQSLPRR50 pKa = 11.84RR51 pKa = 11.84AQNILSLPQRR61 pKa = 11.84PRR63 pKa = 11.84NAQSLPQRR71 pKa = 11.84PQNTQNLPQRR81 pKa = 11.84PRR83 pKa = 11.84NIVNLPRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84NIVSLPGRR101 pKa = 11.84LRR103 pKa = 11.84NAQSLPWRR111 pKa = 11.84PRR113 pKa = 11.84NILNLPQRR121 pKa = 11.84PRR123 pKa = 11.84NAQSLPRR130 pKa = 11.84KK131 pKa = 8.74PRR133 pKa = 11.84NAQSLSRR140 pKa = 11.84RR141 pKa = 11.84PLNTQSLPRR150 pKa = 11.84GPRR153 pKa = 11.84NAQSLPQRR161 pKa = 11.84PWNAQSLPCGSQNAQSLSQRR181 pKa = 11.84PWNAQSLPQGSRR193 pKa = 11.84SILSLPRR200 pKa = 11.84KK201 pKa = 9.26LRR203 pKa = 11.84NAQSLPRR210 pKa = 11.84RR211 pKa = 11.84PQNTQSLQQKK221 pKa = 7.64PQNILSLSQRR231 pKa = 11.84PQNAQNLPRR240 pKa = 11.84RR241 pKa = 11.84PQNILSLPQKK251 pKa = 9.74PRR253 pKa = 11.84NAQSLPRR260 pKa = 11.84RR261 pKa = 11.84PQNTQSLPRR270 pKa = 11.84RR271 pKa = 11.84PQNAQNLPQRR281 pKa = 11.84PQNAQSLPQRR291 pKa = 11.84PQNTQNLQRR300 pKa = 11.84RR301 pKa = 11.84PQNAQNLPQGPQNILSLPQRR321 pKa = 11.84PQNAQSLPP329 pKa = 3.6
Molecular weight: 37.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
18266267 |
31 |
34619 |
533.7 |
59.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.002 ± 0.015 | 2.159 ± 0.011 |
4.79 ± 0.008 | 7.202 ± 0.019 |
3.562 ± 0.009 | 6.589 ± 0.018 |
2.617 ± 0.007 | 4.203 ± 0.012 |
5.763 ± 0.019 | 9.918 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.134 ± 0.006 | 3.509 ± 0.011 |
6.465 ± 0.024 | 4.856 ± 0.013 |
5.796 ± 0.015 | 8.469 ± 0.019 |
5.216 ± 0.01 | 5.965 ± 0.012 |
1.188 ± 0.005 | 2.563 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
