
Barley yellow striate mosaic cytorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
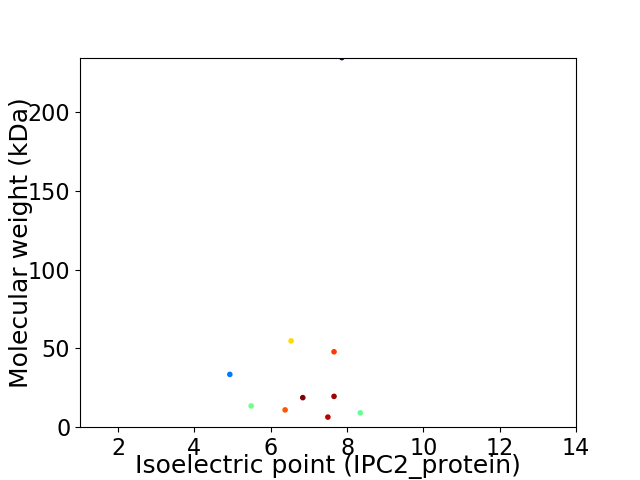
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0C5KH28|A0A0C5KH28_9RHAB M protein OS=Barley yellow striate mosaic cytorhabdovirus OX=1985699 PE=4 SV=1
MM1 pKa = 7.43SSSNAAVTFDD11 pKa = 5.22DD12 pKa = 5.79DD13 pKa = 4.38VPDD16 pKa = 5.73DD17 pKa = 3.95IAPRR21 pKa = 11.84AYY23 pKa = 10.5DD24 pKa = 4.08LDD26 pKa = 4.88LDD28 pKa = 4.85GIFGEE33 pKa = 4.56QDD35 pKa = 3.06VADD38 pKa = 4.73ADD40 pKa = 4.11RR41 pKa = 11.84QAEE44 pKa = 4.08NDD46 pKa = 3.47VEE48 pKa = 4.21EE49 pKa = 4.41QEE51 pKa = 4.94EE52 pKa = 4.46EE53 pKa = 4.22EE54 pKa = 5.15KK55 pKa = 10.36MVPTIKK61 pKa = 10.53GEE63 pKa = 4.02RR64 pKa = 11.84YY65 pKa = 8.37ISGVEE70 pKa = 3.75EE71 pKa = 3.82LAALLKK77 pKa = 10.83DD78 pKa = 3.38HH79 pKa = 7.02ANRR82 pKa = 11.84RR83 pKa = 11.84GMEE86 pKa = 4.03MKK88 pKa = 10.49KK89 pKa = 9.94EE90 pKa = 3.86WVNIMKK96 pKa = 10.4RR97 pKa = 11.84KK98 pKa = 7.26YY99 pKa = 10.37HH100 pKa = 4.93EE101 pKa = 4.89MKK103 pKa = 10.82DD104 pKa = 3.53KK105 pKa = 10.13MLKK108 pKa = 9.62SHH110 pKa = 7.02VEE112 pKa = 3.84LFLLGIQVEE121 pKa = 4.78RR122 pKa = 11.84NTSVDD127 pKa = 3.35KK128 pKa = 11.05DD129 pKa = 4.2FKK131 pKa = 10.87DD132 pKa = 3.06TATRR136 pKa = 11.84LNDD139 pKa = 3.44EE140 pKa = 4.37VNKK143 pKa = 10.31VSGISKK149 pKa = 10.86KK150 pKa = 10.99LMDD153 pKa = 4.45SQAKK157 pKa = 8.53IAKK160 pKa = 10.02DD161 pKa = 3.19VDD163 pKa = 3.55QKK165 pKa = 10.92MKK167 pKa = 10.72EE168 pKa = 3.73LTAYY172 pKa = 8.5CRR174 pKa = 11.84KK175 pKa = 8.25MEE177 pKa = 4.34SMVSEE182 pKa = 4.23VKK184 pKa = 9.99TVVEE188 pKa = 4.35SASRR192 pKa = 11.84PSSIASWAQGEE203 pKa = 4.37EE204 pKa = 4.53KK205 pKa = 9.82NTSEE209 pKa = 5.13DD210 pKa = 3.41YY211 pKa = 11.25NYY213 pKa = 11.32DD214 pKa = 3.38KK215 pKa = 10.44FLSMIGFSANHH226 pKa = 5.95IRR228 pKa = 11.84SSVMKK233 pKa = 8.99KK234 pKa = 9.28CHH236 pKa = 5.38VAITDD241 pKa = 3.15EE242 pKa = 4.73MYY244 pKa = 9.12THH246 pKa = 6.98VISGEE251 pKa = 3.89ADD253 pKa = 3.3SDD255 pKa = 4.3DD256 pKa = 3.49MADD259 pKa = 3.53YY260 pKa = 10.89YY261 pKa = 11.73SRR263 pKa = 11.84ILNYY267 pKa = 10.05ADD269 pKa = 3.79RR270 pKa = 11.84VVKK273 pKa = 10.71GSSKK277 pKa = 10.02EE278 pKa = 3.85EE279 pKa = 3.79RR280 pKa = 11.84TRR282 pKa = 11.84PASSSRR288 pKa = 11.84HH289 pKa = 5.77DD290 pKa = 3.85PYY292 pKa = 11.92GDD294 pKa = 3.36LL295 pKa = 4.72
MM1 pKa = 7.43SSSNAAVTFDD11 pKa = 5.22DD12 pKa = 5.79DD13 pKa = 4.38VPDD16 pKa = 5.73DD17 pKa = 3.95IAPRR21 pKa = 11.84AYY23 pKa = 10.5DD24 pKa = 4.08LDD26 pKa = 4.88LDD28 pKa = 4.85GIFGEE33 pKa = 4.56QDD35 pKa = 3.06VADD38 pKa = 4.73ADD40 pKa = 4.11RR41 pKa = 11.84QAEE44 pKa = 4.08NDD46 pKa = 3.47VEE48 pKa = 4.21EE49 pKa = 4.41QEE51 pKa = 4.94EE52 pKa = 4.46EE53 pKa = 4.22EE54 pKa = 5.15KK55 pKa = 10.36MVPTIKK61 pKa = 10.53GEE63 pKa = 4.02RR64 pKa = 11.84YY65 pKa = 8.37ISGVEE70 pKa = 3.75EE71 pKa = 3.82LAALLKK77 pKa = 10.83DD78 pKa = 3.38HH79 pKa = 7.02ANRR82 pKa = 11.84RR83 pKa = 11.84GMEE86 pKa = 4.03MKK88 pKa = 10.49KK89 pKa = 9.94EE90 pKa = 3.86WVNIMKK96 pKa = 10.4RR97 pKa = 11.84KK98 pKa = 7.26YY99 pKa = 10.37HH100 pKa = 4.93EE101 pKa = 4.89MKK103 pKa = 10.82DD104 pKa = 3.53KK105 pKa = 10.13MLKK108 pKa = 9.62SHH110 pKa = 7.02VEE112 pKa = 3.84LFLLGIQVEE121 pKa = 4.78RR122 pKa = 11.84NTSVDD127 pKa = 3.35KK128 pKa = 11.05DD129 pKa = 4.2FKK131 pKa = 10.87DD132 pKa = 3.06TATRR136 pKa = 11.84LNDD139 pKa = 3.44EE140 pKa = 4.37VNKK143 pKa = 10.31VSGISKK149 pKa = 10.86KK150 pKa = 10.99LMDD153 pKa = 4.45SQAKK157 pKa = 8.53IAKK160 pKa = 10.02DD161 pKa = 3.19VDD163 pKa = 3.55QKK165 pKa = 10.92MKK167 pKa = 10.72EE168 pKa = 3.73LTAYY172 pKa = 8.5CRR174 pKa = 11.84KK175 pKa = 8.25MEE177 pKa = 4.34SMVSEE182 pKa = 4.23VKK184 pKa = 9.99TVVEE188 pKa = 4.35SASRR192 pKa = 11.84PSSIASWAQGEE203 pKa = 4.37EE204 pKa = 4.53KK205 pKa = 9.82NTSEE209 pKa = 5.13DD210 pKa = 3.41YY211 pKa = 11.25NYY213 pKa = 11.32DD214 pKa = 3.38KK215 pKa = 10.44FLSMIGFSANHH226 pKa = 5.95IRR228 pKa = 11.84SSVMKK233 pKa = 8.99KK234 pKa = 9.28CHH236 pKa = 5.38VAITDD241 pKa = 3.15EE242 pKa = 4.73MYY244 pKa = 9.12THH246 pKa = 6.98VISGEE251 pKa = 3.89ADD253 pKa = 3.3SDD255 pKa = 4.3DD256 pKa = 3.49MADD259 pKa = 3.53YY260 pKa = 10.89YY261 pKa = 11.73SRR263 pKa = 11.84ILNYY267 pKa = 10.05ADD269 pKa = 3.79RR270 pKa = 11.84VVKK273 pKa = 10.71GSSKK277 pKa = 10.02EE278 pKa = 3.85EE279 pKa = 3.79RR280 pKa = 11.84TRR282 pKa = 11.84PASSSRR288 pKa = 11.84HH289 pKa = 5.77DD290 pKa = 3.85PYY292 pKa = 11.92GDD294 pKa = 3.36LL295 pKa = 4.72
Molecular weight: 33.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5KQX4|A0A0C5KQX4_9RHAB RNA-directed RNA polymerase L OS=Barley yellow striate mosaic cytorhabdovirus OX=1985699 GN=L PE=3 SV=1
MM1 pKa = 7.55WNSLWLYY8 pKa = 10.65LVDD11 pKa = 4.61GLSQEE16 pKa = 4.31RR17 pKa = 11.84TSSFWATLPFSVQLILAIMILTSLCRR43 pKa = 11.84LKK45 pKa = 10.85VYY47 pKa = 8.02QLKK50 pKa = 10.26YY51 pKa = 10.06FAIISAFQLLAALTSLFRR69 pKa = 11.84EE70 pKa = 4.23LMYY73 pKa = 10.3ATGHH77 pKa = 4.94STT79 pKa = 3.4
MM1 pKa = 7.55WNSLWLYY8 pKa = 10.65LVDD11 pKa = 4.61GLSQEE16 pKa = 4.31RR17 pKa = 11.84TSSFWATLPFSVQLILAIMILTSLCRR43 pKa = 11.84LKK45 pKa = 10.85VYY47 pKa = 8.02QLKK50 pKa = 10.26YY51 pKa = 10.06FAIISAFQLLAALTSLFRR69 pKa = 11.84EE70 pKa = 4.23LMYY73 pKa = 10.3ATGHH77 pKa = 4.94STT79 pKa = 3.4
Molecular weight: 9.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3947 |
51 |
2056 |
394.7 |
44.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.27 ± 0.637 | 1.596 ± 0.466 |
5.65 ± 0.575 | 6.41 ± 0.606 |
3.851 ± 0.32 | 5.599 ± 0.402 |
2.28 ± 0.2 | 7.17 ± 0.427 |
6.866 ± 0.403 | 9.602 ± 0.785 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.344 ± 0.359 | 4.104 ± 0.341 |
3.902 ± 0.578 | 2.711 ± 0.227 |
5.447 ± 0.26 | 8.766 ± 0.403 |
5.726 ± 0.417 | 6.258 ± 0.277 |
1.571 ± 0.253 | 3.876 ± 0.263 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
