
Oceanococcus atlanticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Oceanococcus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
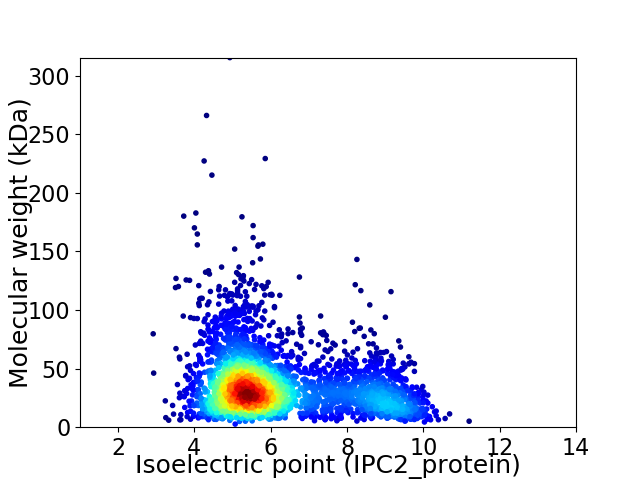
Virtual 2D-PAGE plot for 3284 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y1SCE4|A0A1Y1SCE4_9GAMM LuxR family transcriptional regulator OS=Oceanococcus atlanticus OX=1317117 GN=ATO7_11898 PE=4 SV=1
MM1 pKa = 7.73TIQKK5 pKa = 8.93TGLAVLGLTAAAMVGCGGGGGGGGSSDD32 pKa = 3.87PTPTPTPTPTATPGAGDD49 pKa = 4.08SVCDD53 pKa = 3.79EE54 pKa = 4.75SFVSCSGTNATLSGTIDD71 pKa = 3.25KK72 pKa = 10.89DD73 pKa = 3.72FTLSTDD79 pKa = 4.16FNWFLDD85 pKa = 3.59GFVQVGNGNVEE96 pKa = 3.97ITSNAQANDD105 pKa = 3.05LRR107 pKa = 11.84NNGVEE112 pKa = 4.01LTIPAGTHH120 pKa = 4.03VKK122 pKa = 10.46ALEE125 pKa = 4.13TGVLIVTRR133 pKa = 11.84GNRR136 pKa = 11.84LIANGTAANPITFSSVDD153 pKa = 3.66DD154 pKa = 4.67GFDD157 pKa = 4.19GLGEE161 pKa = 3.94WGGVVLQGFAPQFGAGGTGNCFGDD185 pKa = 3.54NNYY188 pKa = 10.34CNVAGEE194 pKa = 4.55GGSDD198 pKa = 3.14VAFYY202 pKa = 10.6GGKK205 pKa = 10.14YY206 pKa = 9.24PADD209 pKa = 3.35NSGSIRR215 pKa = 11.84YY216 pKa = 8.84VRR218 pKa = 11.84IAEE221 pKa = 4.45GGLVAGPNNEE231 pKa = 4.11VNGLTLQGVGHH242 pKa = 7.2ATTIEE247 pKa = 4.08YY248 pKa = 10.21VQVHH252 pKa = 6.33NNLDD256 pKa = 4.23DD257 pKa = 4.11GVEE260 pKa = 4.1WFGGTVNAKK269 pKa = 10.18YY270 pKa = 10.37LVLTGNDD277 pKa = 4.58DD278 pKa = 4.52DD279 pKa = 7.02DD280 pKa = 4.77IDD282 pKa = 4.24YY283 pKa = 11.56DD284 pKa = 3.74EE285 pKa = 5.51GYY287 pKa = 10.28KK288 pKa = 11.09GNIQFAIVKK297 pKa = 9.74KK298 pKa = 10.32SSNDD302 pKa = 3.15APQGSNDD309 pKa = 3.25PRR311 pKa = 11.84GIEE314 pKa = 4.23ANSSDD319 pKa = 4.26EE320 pKa = 4.74DD321 pKa = 3.88FVPQTTASLANILLIGNTVSANEE344 pKa = 3.65PGMRR348 pKa = 11.84LRR350 pKa = 11.84GALTTSIFNTALVGWDD366 pKa = 3.71KK367 pKa = 11.61GCIRR371 pKa = 11.84IDD373 pKa = 4.42DD374 pKa = 4.66ADD376 pKa = 3.99TNADD380 pKa = 3.57DD381 pKa = 5.35VADD384 pKa = 3.67QFSNVSLEE392 pKa = 4.2NVFGDD397 pKa = 4.21CAGGFYY403 pKa = 8.92THH405 pKa = 7.11EE406 pKa = 5.06DD407 pKa = 3.52ADD409 pKa = 4.05STAGGVGSQTISLDD423 pKa = 3.07DD424 pKa = 4.2AYY426 pKa = 10.73AINEE430 pKa = 4.19GFASLASATNIVAVDD445 pKa = 3.69NGSGFAFEE453 pKa = 4.22TTDD456 pKa = 3.21YY457 pKa = 11.1VGAVEE462 pKa = 5.33PGTSMGDD469 pKa = 3.13AWWAGWTIPGSLSDD483 pKa = 4.44ADD485 pKa = 4.12EE486 pKa = 4.95APASASFVSCNDD498 pKa = 2.51STMICEE504 pKa = 4.08VTGTIDD510 pKa = 3.22EE511 pKa = 5.47DD512 pKa = 4.25YY513 pKa = 10.38TFVAGYY519 pKa = 7.95TWVLNGLVKK528 pKa = 10.94VGDD531 pKa = 3.99GNVEE535 pKa = 4.06INSEE539 pKa = 4.12SQADD543 pKa = 3.76QIKK546 pKa = 10.8ADD548 pKa = 3.79GVTLTIRR555 pKa = 11.84PGVDD559 pKa = 2.95VKK561 pKa = 10.78GTEE564 pKa = 3.98TGVLVVTRR572 pKa = 11.84GSKK575 pKa = 10.73LIANGSASAPITFSSLDD592 pKa = 3.57EE593 pKa = 4.55NFDD596 pKa = 4.45GLGEE600 pKa = 4.03WGGVVLQGFAPQFGAGNTGNCHH622 pKa = 6.16GANSYY627 pKa = 11.29CNVAGEE633 pKa = 4.52GGGGVAFYY641 pKa = 10.96GGDD644 pKa = 3.65DD645 pKa = 3.66PADD648 pKa = 3.39NSGSLRR654 pKa = 11.84YY655 pKa = 10.08VRR657 pKa = 11.84IAEE660 pKa = 4.48GGLVAGPNNEE670 pKa = 4.11VNGLTLQGVGYY681 pKa = 7.25MTSLDD686 pKa = 3.98YY687 pKa = 11.1IQVHH691 pKa = 6.17NNLDD695 pKa = 4.34DD696 pKa = 4.11GVEE699 pKa = 4.1WFGGTVNAKK708 pKa = 10.11HH709 pKa = 6.32LVLTGNDD716 pKa = 3.82DD717 pKa = 4.09DD718 pKa = 7.3DD719 pKa = 5.42IDD721 pKa = 4.24FDD723 pKa = 4.14EE724 pKa = 5.78GYY726 pKa = 10.36KK727 pKa = 11.14GNIQYY732 pKa = 10.93AIVIKK737 pKa = 10.5SDD739 pKa = 2.86NATPQGSNDD748 pKa = 3.19PRR750 pKa = 11.84GIEE753 pKa = 4.23ANSSDD758 pKa = 4.19EE759 pKa = 5.17DD760 pKa = 4.07YY761 pKa = 11.56VPQTQATLANILLVGNTVSANEE783 pKa = 3.65PGMRR787 pKa = 11.84LRR789 pKa = 11.84GALTVSIFNTALTGWDD805 pKa = 3.74EE806 pKa = 4.12GCIRR810 pKa = 11.84IDD812 pKa = 4.59DD813 pKa = 4.52ADD815 pKa = 4.12TNGDD819 pKa = 3.62DD820 pKa = 4.06TNDD823 pKa = 3.39AFSDD827 pKa = 3.75VTLVNILGNCSGGLYY842 pKa = 6.85THH844 pKa = 7.52RR845 pKa = 11.84DD846 pKa = 3.1ADD848 pKa = 3.67SQTNYY853 pKa = 10.05VSNAVNLNQALVVTGAAAQVSAPSITAVANGSGFSFDD890 pKa = 3.15EE891 pKa = 4.15TNYY894 pKa = 10.1IGAVAPGTNVADD906 pKa = 3.78AWWNGWTIPNSVSVDD921 pKa = 3.17
MM1 pKa = 7.73TIQKK5 pKa = 8.93TGLAVLGLTAAAMVGCGGGGGGGGSSDD32 pKa = 3.87PTPTPTPTPTATPGAGDD49 pKa = 4.08SVCDD53 pKa = 3.79EE54 pKa = 4.75SFVSCSGTNATLSGTIDD71 pKa = 3.25KK72 pKa = 10.89DD73 pKa = 3.72FTLSTDD79 pKa = 4.16FNWFLDD85 pKa = 3.59GFVQVGNGNVEE96 pKa = 3.97ITSNAQANDD105 pKa = 3.05LRR107 pKa = 11.84NNGVEE112 pKa = 4.01LTIPAGTHH120 pKa = 4.03VKK122 pKa = 10.46ALEE125 pKa = 4.13TGVLIVTRR133 pKa = 11.84GNRR136 pKa = 11.84LIANGTAANPITFSSVDD153 pKa = 3.66DD154 pKa = 4.67GFDD157 pKa = 4.19GLGEE161 pKa = 3.94WGGVVLQGFAPQFGAGGTGNCFGDD185 pKa = 3.54NNYY188 pKa = 10.34CNVAGEE194 pKa = 4.55GGSDD198 pKa = 3.14VAFYY202 pKa = 10.6GGKK205 pKa = 10.14YY206 pKa = 9.24PADD209 pKa = 3.35NSGSIRR215 pKa = 11.84YY216 pKa = 8.84VRR218 pKa = 11.84IAEE221 pKa = 4.45GGLVAGPNNEE231 pKa = 4.11VNGLTLQGVGHH242 pKa = 7.2ATTIEE247 pKa = 4.08YY248 pKa = 10.21VQVHH252 pKa = 6.33NNLDD256 pKa = 4.23DD257 pKa = 4.11GVEE260 pKa = 4.1WFGGTVNAKK269 pKa = 10.18YY270 pKa = 10.37LVLTGNDD277 pKa = 4.58DD278 pKa = 4.52DD279 pKa = 7.02DD280 pKa = 4.77IDD282 pKa = 4.24YY283 pKa = 11.56DD284 pKa = 3.74EE285 pKa = 5.51GYY287 pKa = 10.28KK288 pKa = 11.09GNIQFAIVKK297 pKa = 9.74KK298 pKa = 10.32SSNDD302 pKa = 3.15APQGSNDD309 pKa = 3.25PRR311 pKa = 11.84GIEE314 pKa = 4.23ANSSDD319 pKa = 4.26EE320 pKa = 4.74DD321 pKa = 3.88FVPQTTASLANILLIGNTVSANEE344 pKa = 3.65PGMRR348 pKa = 11.84LRR350 pKa = 11.84GALTTSIFNTALVGWDD366 pKa = 3.71KK367 pKa = 11.61GCIRR371 pKa = 11.84IDD373 pKa = 4.42DD374 pKa = 4.66ADD376 pKa = 3.99TNADD380 pKa = 3.57DD381 pKa = 5.35VADD384 pKa = 3.67QFSNVSLEE392 pKa = 4.2NVFGDD397 pKa = 4.21CAGGFYY403 pKa = 8.92THH405 pKa = 7.11EE406 pKa = 5.06DD407 pKa = 3.52ADD409 pKa = 4.05STAGGVGSQTISLDD423 pKa = 3.07DD424 pKa = 4.2AYY426 pKa = 10.73AINEE430 pKa = 4.19GFASLASATNIVAVDD445 pKa = 3.69NGSGFAFEE453 pKa = 4.22TTDD456 pKa = 3.21YY457 pKa = 11.1VGAVEE462 pKa = 5.33PGTSMGDD469 pKa = 3.13AWWAGWTIPGSLSDD483 pKa = 4.44ADD485 pKa = 4.12EE486 pKa = 4.95APASASFVSCNDD498 pKa = 2.51STMICEE504 pKa = 4.08VTGTIDD510 pKa = 3.22EE511 pKa = 5.47DD512 pKa = 4.25YY513 pKa = 10.38TFVAGYY519 pKa = 7.95TWVLNGLVKK528 pKa = 10.94VGDD531 pKa = 3.99GNVEE535 pKa = 4.06INSEE539 pKa = 4.12SQADD543 pKa = 3.76QIKK546 pKa = 10.8ADD548 pKa = 3.79GVTLTIRR555 pKa = 11.84PGVDD559 pKa = 2.95VKK561 pKa = 10.78GTEE564 pKa = 3.98TGVLVVTRR572 pKa = 11.84GSKK575 pKa = 10.73LIANGSASAPITFSSLDD592 pKa = 3.57EE593 pKa = 4.55NFDD596 pKa = 4.45GLGEE600 pKa = 4.03WGGVVLQGFAPQFGAGNTGNCHH622 pKa = 6.16GANSYY627 pKa = 11.29CNVAGEE633 pKa = 4.52GGGGVAFYY641 pKa = 10.96GGDD644 pKa = 3.65DD645 pKa = 3.66PADD648 pKa = 3.39NSGSLRR654 pKa = 11.84YY655 pKa = 10.08VRR657 pKa = 11.84IAEE660 pKa = 4.48GGLVAGPNNEE670 pKa = 4.11VNGLTLQGVGYY681 pKa = 7.25MTSLDD686 pKa = 3.98YY687 pKa = 11.1IQVHH691 pKa = 6.17NNLDD695 pKa = 4.34DD696 pKa = 4.11GVEE699 pKa = 4.1WFGGTVNAKK708 pKa = 10.11HH709 pKa = 6.32LVLTGNDD716 pKa = 3.82DD717 pKa = 4.09DD718 pKa = 7.3DD719 pKa = 5.42IDD721 pKa = 4.24FDD723 pKa = 4.14EE724 pKa = 5.78GYY726 pKa = 10.36KK727 pKa = 11.14GNIQYY732 pKa = 10.93AIVIKK737 pKa = 10.5SDD739 pKa = 2.86NATPQGSNDD748 pKa = 3.19PRR750 pKa = 11.84GIEE753 pKa = 4.23ANSSDD758 pKa = 4.19EE759 pKa = 5.17DD760 pKa = 4.07YY761 pKa = 11.56VPQTQATLANILLVGNTVSANEE783 pKa = 3.65PGMRR787 pKa = 11.84LRR789 pKa = 11.84GALTVSIFNTALTGWDD805 pKa = 3.74EE806 pKa = 4.12GCIRR810 pKa = 11.84IDD812 pKa = 4.59DD813 pKa = 4.52ADD815 pKa = 4.12TNGDD819 pKa = 3.62DD820 pKa = 4.06TNDD823 pKa = 3.39AFSDD827 pKa = 3.75VTLVNILGNCSGGLYY842 pKa = 6.85THH844 pKa = 7.52RR845 pKa = 11.84DD846 pKa = 3.1ADD848 pKa = 3.67SQTNYY853 pKa = 10.05VSNAVNLNQALVVTGAAAQVSAPSITAVANGSGFSFDD890 pKa = 3.15EE891 pKa = 4.15TNYY894 pKa = 10.1IGAVAPGTNVADD906 pKa = 3.78AWWNGWTIPNSVSVDD921 pKa = 3.17
Molecular weight: 94.81 kDa
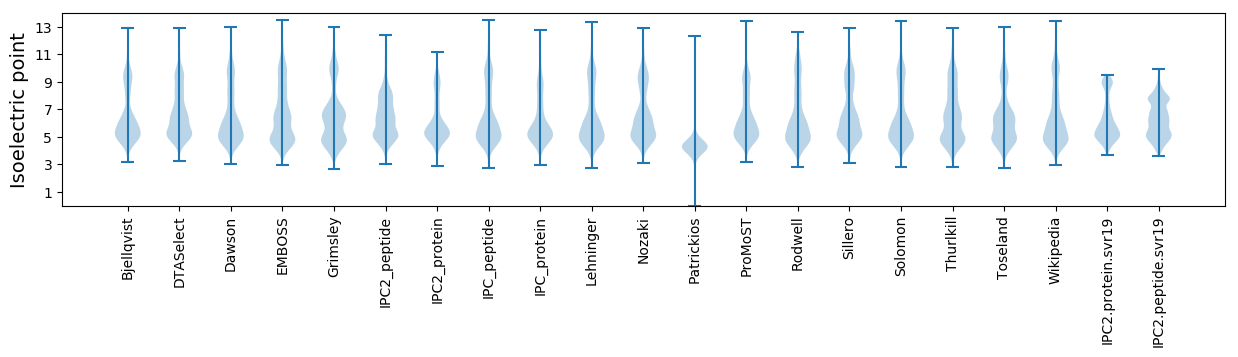
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1SIR0|A0A1Y1SIR0_9GAMM Uncharacterized protein OS=Oceanococcus atlanticus OX=1317117 GN=ATO7_04910 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.13GGRR28 pKa = 11.84RR29 pKa = 11.84VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.85GRR39 pKa = 11.84HH40 pKa = 5.12SLTVV44 pKa = 3.06
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.26RR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.13GGRR28 pKa = 11.84RR29 pKa = 11.84VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.85GRR39 pKa = 11.84HH40 pKa = 5.12SLTVV44 pKa = 3.06
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1129603 |
27 |
2975 |
344.0 |
37.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.645 ± 0.06 | 1.003 ± 0.015 |
6.12 ± 0.036 | 5.63 ± 0.036 |
3.495 ± 0.022 | 7.908 ± 0.041 |
2.378 ± 0.021 | 4.671 ± 0.032 |
2.822 ± 0.034 | 10.96 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.364 ± 0.022 | 2.958 ± 0.026 |
4.908 ± 0.026 | 4.576 ± 0.029 |
6.897 ± 0.043 | 5.877 ± 0.035 |
4.729 ± 0.031 | 7.059 ± 0.038 |
1.492 ± 0.017 | 2.507 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
