
Bat polyomavirus 6a
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Acerodon celebensis polyomavirus 2
Average proteome isoelectric point is 5.62
Get precalculated fractions of proteins
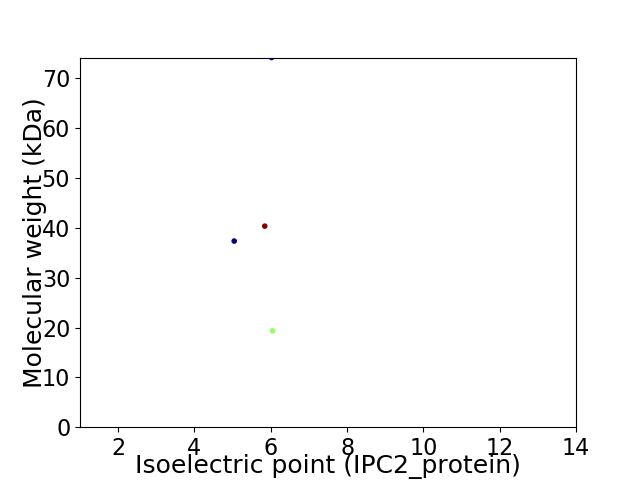
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5ZYL6|A0A0D5ZYL6_9POLY Small t antigen OS=Bat polyomavirus 6a OX=1623685 PE=4 SV=1
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.31VFDD13 pKa = 4.55LAATTGLSVDD23 pKa = 3.57SFLTGEE29 pKa = 4.4AFTTAEE35 pKa = 4.17VLQSHH40 pKa = 7.13IANLVTYY47 pKa = 9.75GGLSEE52 pKa = 5.27AEE54 pKa = 3.8ALAAAEE60 pKa = 4.48VSQEE64 pKa = 4.02AYY66 pKa = 10.37SALTSLTSNFPRR78 pKa = 11.84AFAALAGTEE87 pKa = 3.82ALAVGSLVIGSATAAALAPYY107 pKa = 8.79TFDD110 pKa = 3.22YY111 pKa = 8.75STPIANLNVDD121 pKa = 3.4MALQVWQPNWDD132 pKa = 4.25DD133 pKa = 3.23IFFPGVVPFARR144 pKa = 11.84IVNYY148 pKa = 9.84IDD150 pKa = 3.56PANWAAYY157 pKa = 9.76LYY159 pKa = 9.93QAVGRR164 pKa = 11.84YY165 pKa = 7.22FWEE168 pKa = 4.19TAQRR172 pKa = 11.84TGQHH176 pKa = 6.24LIEE179 pKa = 5.69HH180 pKa = 6.53EE181 pKa = 4.15VRR183 pKa = 11.84EE184 pKa = 4.32VARR187 pKa = 11.84EE188 pKa = 3.76VGQRR192 pKa = 11.84TVQSVSEE199 pKa = 3.87TLARR203 pKa = 11.84YY204 pKa = 9.22FEE206 pKa = 4.06NARR209 pKa = 11.84WAVSHH214 pKa = 6.78LSSNVYY220 pKa = 10.4SGLQNYY226 pKa = 7.06YY227 pKa = 10.03TEE229 pKa = 5.13LGPLRR234 pKa = 11.84PHH236 pKa = 5.8QVRR239 pKa = 11.84AVNRR243 pKa = 11.84RR244 pKa = 11.84LGRR247 pKa = 11.84EE248 pKa = 3.47LPTRR252 pKa = 11.84YY253 pKa = 9.72NLEE256 pKa = 3.98TPRR259 pKa = 11.84PQDD262 pKa = 3.09KK263 pKa = 10.96ASAQYY268 pKa = 8.41VTKK271 pKa = 10.7SDD273 pKa = 3.74APGGAHH279 pKa = 6.4QRR281 pKa = 11.84VTPDD285 pKa = 2.33WMLPLILGLYY295 pKa = 10.55GDD297 pKa = 4.68ITPSWEE303 pKa = 3.81ATLEE307 pKa = 4.05DD308 pKa = 3.98LEE310 pKa = 4.45EE311 pKa = 4.39EE312 pKa = 4.35EE313 pKa = 4.89EE314 pKa = 4.29EE315 pKa = 4.78EE316 pKa = 4.66EE317 pKa = 4.79NGPKK321 pKa = 9.81KK322 pKa = 10.36KK323 pKa = 10.22KK324 pKa = 9.31PRR326 pKa = 11.84RR327 pKa = 11.84GKK329 pKa = 10.15KK330 pKa = 5.74QTKK333 pKa = 9.91ANSNSSAA340 pKa = 3.2
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.31VFDD13 pKa = 4.55LAATTGLSVDD23 pKa = 3.57SFLTGEE29 pKa = 4.4AFTTAEE35 pKa = 4.17VLQSHH40 pKa = 7.13IANLVTYY47 pKa = 9.75GGLSEE52 pKa = 5.27AEE54 pKa = 3.8ALAAAEE60 pKa = 4.48VSQEE64 pKa = 4.02AYY66 pKa = 10.37SALTSLTSNFPRR78 pKa = 11.84AFAALAGTEE87 pKa = 3.82ALAVGSLVIGSATAAALAPYY107 pKa = 8.79TFDD110 pKa = 3.22YY111 pKa = 8.75STPIANLNVDD121 pKa = 3.4MALQVWQPNWDD132 pKa = 4.25DD133 pKa = 3.23IFFPGVVPFARR144 pKa = 11.84IVNYY148 pKa = 9.84IDD150 pKa = 3.56PANWAAYY157 pKa = 9.76LYY159 pKa = 9.93QAVGRR164 pKa = 11.84YY165 pKa = 7.22FWEE168 pKa = 4.19TAQRR172 pKa = 11.84TGQHH176 pKa = 6.24LIEE179 pKa = 5.69HH180 pKa = 6.53EE181 pKa = 4.15VRR183 pKa = 11.84EE184 pKa = 4.32VARR187 pKa = 11.84EE188 pKa = 3.76VGQRR192 pKa = 11.84TVQSVSEE199 pKa = 3.87TLARR203 pKa = 11.84YY204 pKa = 9.22FEE206 pKa = 4.06NARR209 pKa = 11.84WAVSHH214 pKa = 6.78LSSNVYY220 pKa = 10.4SGLQNYY226 pKa = 7.06YY227 pKa = 10.03TEE229 pKa = 5.13LGPLRR234 pKa = 11.84PHH236 pKa = 5.8QVRR239 pKa = 11.84AVNRR243 pKa = 11.84RR244 pKa = 11.84LGRR247 pKa = 11.84EE248 pKa = 3.47LPTRR252 pKa = 11.84YY253 pKa = 9.72NLEE256 pKa = 3.98TPRR259 pKa = 11.84PQDD262 pKa = 3.09KK263 pKa = 10.96ASAQYY268 pKa = 8.41VTKK271 pKa = 10.7SDD273 pKa = 3.74APGGAHH279 pKa = 6.4QRR281 pKa = 11.84VTPDD285 pKa = 2.33WMLPLILGLYY295 pKa = 10.55GDD297 pKa = 4.68ITPSWEE303 pKa = 3.81ATLEE307 pKa = 4.05DD308 pKa = 3.98LEE310 pKa = 4.45EE311 pKa = 4.39EE312 pKa = 4.35EE313 pKa = 4.89EE314 pKa = 4.29EE315 pKa = 4.78EE316 pKa = 4.66EE317 pKa = 4.79NGPKK321 pKa = 9.81KK322 pKa = 10.36KK323 pKa = 10.22KK324 pKa = 9.31PRR326 pKa = 11.84RR327 pKa = 11.84GKK329 pKa = 10.15KK330 pKa = 5.74QTKK333 pKa = 9.91ANSNSSAA340 pKa = 3.2
Molecular weight: 37.34 kDa
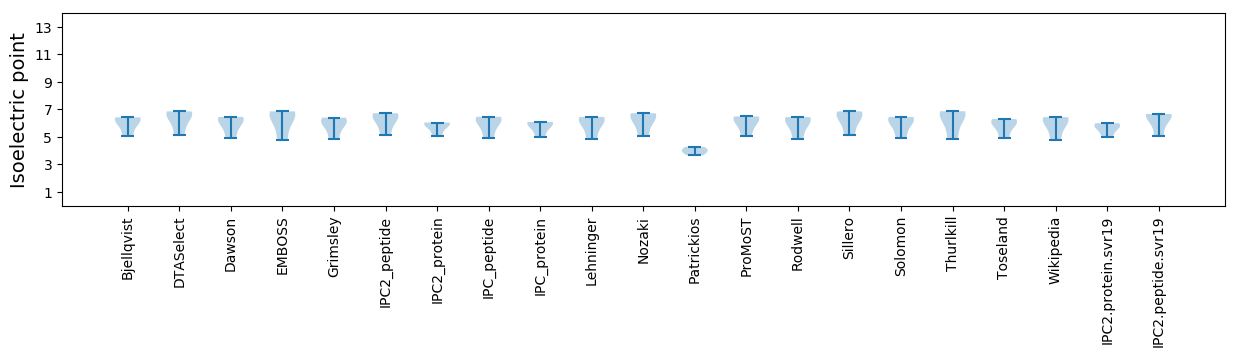
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5ZZ24|A0A0D5ZZ24_9POLY Large T antigen OS=Bat polyomavirus 6a OX=1623685 PE=4 SV=1
MM1 pKa = 7.66DD2 pKa = 4.87HH3 pKa = 7.31ALTRR7 pKa = 11.84EE8 pKa = 3.84EE9 pKa = 4.5SVRR12 pKa = 11.84LMQLLKK18 pKa = 11.02LPMEE22 pKa = 4.28QYY24 pKa = 11.86GNFSLMRR31 pKa = 11.84KK32 pKa = 9.53AFLRR36 pKa = 11.84EE37 pKa = 4.23CKK39 pKa = 10.0NLHH42 pKa = 6.42PDD44 pKa = 2.95KK45 pKa = 11.24GGNPEE50 pKa = 4.08LAKK53 pKa = 10.59EE54 pKa = 4.41LITLYY59 pKa = 10.67RR60 pKa = 11.84KK61 pKa = 10.23LEE63 pKa = 4.16EE64 pKa = 3.86QAPPYY69 pKa = 7.55GTPEE73 pKa = 3.51WDD75 pKa = 3.28RR76 pKa = 11.84WWEE79 pKa = 3.84EE80 pKa = 3.8FNDD83 pKa = 4.17LFCHH87 pKa = 5.08EE88 pKa = 4.52TFDD91 pKa = 4.9RR92 pKa = 11.84SDD94 pKa = 3.67DD95 pKa = 3.71EE96 pKa = 4.29EE97 pKa = 4.28EE98 pKa = 4.03QRR100 pKa = 11.84KK101 pKa = 8.95RR102 pKa = 11.84PASPEE107 pKa = 4.01SEE109 pKa = 4.47PEE111 pKa = 4.14CSQATPPKK119 pKa = 9.81KK120 pKa = 9.94KK121 pKa = 10.54KK122 pKa = 9.62KK123 pKa = 9.8DD124 pKa = 3.51HH125 pKa = 6.69PKK127 pKa = 11.04DD128 pKa = 3.55MPEE131 pKa = 4.59DD132 pKa = 3.57LQQFLSNAILSNKK145 pKa = 7.54TVSSFLIFTTAEE157 pKa = 4.16KK158 pKa = 9.98STLLYY163 pKa = 10.79GKK165 pKa = 10.29LRR167 pKa = 11.84DD168 pKa = 3.55RR169 pKa = 11.84FKK171 pKa = 10.01ATFVSRR177 pKa = 11.84HH178 pKa = 4.94KK179 pKa = 10.58QQGSDD184 pKa = 3.11GEE186 pKa = 4.39AFVFLITPSRR196 pKa = 11.84HH197 pKa = 4.53RR198 pKa = 11.84VSAINNFCNSLCSVSFIIVKK218 pKa = 9.98GVIKK222 pKa = 10.19EE223 pKa = 4.03YY224 pKa = 10.8NLYY227 pKa = 10.39VHH229 pKa = 6.95LCVEE233 pKa = 5.04PYY235 pKa = 10.91VKK237 pKa = 10.38LQEE240 pKa = 4.7SIPGGLSRR248 pKa = 11.84DD249 pKa = 3.68FFDD252 pKa = 4.47TPEE255 pKa = 3.82EE256 pKa = 3.88AAKK259 pKa = 10.37NVSWKK264 pKa = 9.92MIAEE268 pKa = 4.53FALDD272 pKa = 4.64LMCDD276 pKa = 4.07DD277 pKa = 4.91IFLLMGMYY285 pKa = 10.12KK286 pKa = 10.2EE287 pKa = 4.19FAEE290 pKa = 5.05DD291 pKa = 3.41PKK293 pKa = 11.48SCAKK297 pKa = 10.55CEE299 pKa = 3.77AKK301 pKa = 10.27IISDD305 pKa = 3.76HH306 pKa = 5.72FQNHH310 pKa = 4.18FHH312 pKa = 6.88HH313 pKa = 6.85YY314 pKa = 9.89EE315 pKa = 3.87NALLFLDD322 pKa = 4.82CRR324 pKa = 11.84NQKK327 pKa = 10.21AICQQAVDD335 pKa = 3.89GVIAHH340 pKa = 6.58RR341 pKa = 11.84RR342 pKa = 11.84VQTAQLSRR350 pKa = 11.84EE351 pKa = 3.93QQLAEE356 pKa = 3.92RR357 pKa = 11.84FKK359 pKa = 11.32KK360 pKa = 10.61LFEE363 pKa = 4.26KK364 pKa = 10.39MEE366 pKa = 4.3NLFGARR372 pKa = 11.84SKK374 pKa = 9.88VTLRR378 pKa = 11.84TYY380 pKa = 9.45MAGVAWFDD388 pKa = 3.69SLFPDD393 pKa = 4.28KK394 pKa = 11.0SFKK397 pKa = 10.6EE398 pKa = 3.84LLLDD402 pKa = 4.37FLEE405 pKa = 4.74CMVNNVPKK413 pKa = 10.08KK414 pKa = 10.23RR415 pKa = 11.84YY416 pKa = 8.33WFFTGPVNTGKK427 pKa = 7.57TTLAAALLDD436 pKa = 3.8LCGGKK441 pKa = 9.46SLNVNMPFEE450 pKa = 4.38KK451 pKa = 10.86LNFEE455 pKa = 4.68LGVAIDD461 pKa = 3.38QFMVIFEE468 pKa = 4.27DD469 pKa = 3.55VKK471 pKa = 11.36GQVSEE476 pKa = 4.31NKK478 pKa = 9.82NLPTGQGINNLDD490 pKa = 3.79HH491 pKa = 7.04LRR493 pKa = 11.84DD494 pKa = 3.89YY495 pKa = 11.54LDD497 pKa = 3.82GAVKK501 pKa = 10.66VNLEE505 pKa = 4.2KK506 pKa = 10.85KK507 pKa = 9.75HH508 pKa = 6.21LNKK511 pKa = 10.01KK512 pKa = 8.41SQIFPPGIVTANEE525 pKa = 3.9YY526 pKa = 10.32KK527 pKa = 10.82VPFTLRR533 pKa = 11.84VRR535 pKa = 11.84FCKK538 pKa = 9.96TLTFQFRR545 pKa = 11.84KK546 pKa = 10.01NLYY549 pKa = 8.98LSLRR553 pKa = 11.84KK554 pKa = 8.42TEE556 pKa = 4.53DD557 pKa = 2.94LGTYY561 pKa = 9.68RR562 pKa = 11.84VLQNGICLLLFLVYY576 pKa = 10.18HH577 pKa = 6.27CEE579 pKa = 3.76IEE581 pKa = 4.75DD582 pKa = 4.44FSEE585 pKa = 4.43CLHH588 pKa = 6.1KK589 pKa = 9.58TVEE592 pKa = 3.95QWKK595 pKa = 9.43EE596 pKa = 3.82RR597 pKa = 11.84ISQEE601 pKa = 3.86VSLGDD606 pKa = 3.63YY607 pKa = 11.19LDD609 pKa = 4.2FKK611 pKa = 11.84SNIQKK616 pKa = 10.66GNYY619 pKa = 8.55ILQTTEE625 pKa = 3.66SQGPSTQEE633 pKa = 3.91CEE635 pKa = 4.25TEE637 pKa = 4.27DD638 pKa = 3.99FVNN641 pKa = 3.91
MM1 pKa = 7.66DD2 pKa = 4.87HH3 pKa = 7.31ALTRR7 pKa = 11.84EE8 pKa = 3.84EE9 pKa = 4.5SVRR12 pKa = 11.84LMQLLKK18 pKa = 11.02LPMEE22 pKa = 4.28QYY24 pKa = 11.86GNFSLMRR31 pKa = 11.84KK32 pKa = 9.53AFLRR36 pKa = 11.84EE37 pKa = 4.23CKK39 pKa = 10.0NLHH42 pKa = 6.42PDD44 pKa = 2.95KK45 pKa = 11.24GGNPEE50 pKa = 4.08LAKK53 pKa = 10.59EE54 pKa = 4.41LITLYY59 pKa = 10.67RR60 pKa = 11.84KK61 pKa = 10.23LEE63 pKa = 4.16EE64 pKa = 3.86QAPPYY69 pKa = 7.55GTPEE73 pKa = 3.51WDD75 pKa = 3.28RR76 pKa = 11.84WWEE79 pKa = 3.84EE80 pKa = 3.8FNDD83 pKa = 4.17LFCHH87 pKa = 5.08EE88 pKa = 4.52TFDD91 pKa = 4.9RR92 pKa = 11.84SDD94 pKa = 3.67DD95 pKa = 3.71EE96 pKa = 4.29EE97 pKa = 4.28EE98 pKa = 4.03QRR100 pKa = 11.84KK101 pKa = 8.95RR102 pKa = 11.84PASPEE107 pKa = 4.01SEE109 pKa = 4.47PEE111 pKa = 4.14CSQATPPKK119 pKa = 9.81KK120 pKa = 9.94KK121 pKa = 10.54KK122 pKa = 9.62KK123 pKa = 9.8DD124 pKa = 3.51HH125 pKa = 6.69PKK127 pKa = 11.04DD128 pKa = 3.55MPEE131 pKa = 4.59DD132 pKa = 3.57LQQFLSNAILSNKK145 pKa = 7.54TVSSFLIFTTAEE157 pKa = 4.16KK158 pKa = 9.98STLLYY163 pKa = 10.79GKK165 pKa = 10.29LRR167 pKa = 11.84DD168 pKa = 3.55RR169 pKa = 11.84FKK171 pKa = 10.01ATFVSRR177 pKa = 11.84HH178 pKa = 4.94KK179 pKa = 10.58QQGSDD184 pKa = 3.11GEE186 pKa = 4.39AFVFLITPSRR196 pKa = 11.84HH197 pKa = 4.53RR198 pKa = 11.84VSAINNFCNSLCSVSFIIVKK218 pKa = 9.98GVIKK222 pKa = 10.19EE223 pKa = 4.03YY224 pKa = 10.8NLYY227 pKa = 10.39VHH229 pKa = 6.95LCVEE233 pKa = 5.04PYY235 pKa = 10.91VKK237 pKa = 10.38LQEE240 pKa = 4.7SIPGGLSRR248 pKa = 11.84DD249 pKa = 3.68FFDD252 pKa = 4.47TPEE255 pKa = 3.82EE256 pKa = 3.88AAKK259 pKa = 10.37NVSWKK264 pKa = 9.92MIAEE268 pKa = 4.53FALDD272 pKa = 4.64LMCDD276 pKa = 4.07DD277 pKa = 4.91IFLLMGMYY285 pKa = 10.12KK286 pKa = 10.2EE287 pKa = 4.19FAEE290 pKa = 5.05DD291 pKa = 3.41PKK293 pKa = 11.48SCAKK297 pKa = 10.55CEE299 pKa = 3.77AKK301 pKa = 10.27IISDD305 pKa = 3.76HH306 pKa = 5.72FQNHH310 pKa = 4.18FHH312 pKa = 6.88HH313 pKa = 6.85YY314 pKa = 9.89EE315 pKa = 3.87NALLFLDD322 pKa = 4.82CRR324 pKa = 11.84NQKK327 pKa = 10.21AICQQAVDD335 pKa = 3.89GVIAHH340 pKa = 6.58RR341 pKa = 11.84RR342 pKa = 11.84VQTAQLSRR350 pKa = 11.84EE351 pKa = 3.93QQLAEE356 pKa = 3.92RR357 pKa = 11.84FKK359 pKa = 11.32KK360 pKa = 10.61LFEE363 pKa = 4.26KK364 pKa = 10.39MEE366 pKa = 4.3NLFGARR372 pKa = 11.84SKK374 pKa = 9.88VTLRR378 pKa = 11.84TYY380 pKa = 9.45MAGVAWFDD388 pKa = 3.69SLFPDD393 pKa = 4.28KK394 pKa = 11.0SFKK397 pKa = 10.6EE398 pKa = 3.84LLLDD402 pKa = 4.37FLEE405 pKa = 4.74CMVNNVPKK413 pKa = 10.08KK414 pKa = 10.23RR415 pKa = 11.84YY416 pKa = 8.33WFFTGPVNTGKK427 pKa = 7.57TTLAAALLDD436 pKa = 3.8LCGGKK441 pKa = 9.46SLNVNMPFEE450 pKa = 4.38KK451 pKa = 10.86LNFEE455 pKa = 4.68LGVAIDD461 pKa = 3.38QFMVIFEE468 pKa = 4.27DD469 pKa = 3.55VKK471 pKa = 11.36GQVSEE476 pKa = 4.31NKK478 pKa = 9.82NLPTGQGINNLDD490 pKa = 3.79HH491 pKa = 7.04LRR493 pKa = 11.84DD494 pKa = 3.89YY495 pKa = 11.54LDD497 pKa = 3.82GAVKK501 pKa = 10.66VNLEE505 pKa = 4.2KK506 pKa = 10.85KK507 pKa = 9.75HH508 pKa = 6.21LNKK511 pKa = 10.01KK512 pKa = 8.41SQIFPPGIVTANEE525 pKa = 3.9YY526 pKa = 10.32KK527 pKa = 10.82VPFTLRR533 pKa = 11.84VRR535 pKa = 11.84FCKK538 pKa = 9.96TLTFQFRR545 pKa = 11.84KK546 pKa = 10.01NLYY549 pKa = 8.98LSLRR553 pKa = 11.84KK554 pKa = 8.42TEE556 pKa = 4.53DD557 pKa = 2.94LGTYY561 pKa = 9.68RR562 pKa = 11.84VLQNGICLLLFLVYY576 pKa = 10.18HH577 pKa = 6.27CEE579 pKa = 3.76IEE581 pKa = 4.75DD582 pKa = 4.44FSEE585 pKa = 4.43CLHH588 pKa = 6.1KK589 pKa = 9.58TVEE592 pKa = 3.95QWKK595 pKa = 9.43EE596 pKa = 3.82RR597 pKa = 11.84ISQEE601 pKa = 3.86VSLGDD606 pKa = 3.63YY607 pKa = 11.19LDD609 pKa = 4.2FKK611 pKa = 11.84SNIQKK616 pKa = 10.66GNYY619 pKa = 8.55ILQTTEE625 pKa = 3.66SQGPSTQEE633 pKa = 3.91CEE635 pKa = 4.25TEE637 pKa = 4.27DD638 pKa = 3.99FVNN641 pKa = 3.91
Molecular weight: 74.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1509 |
162 |
641 |
377.3 |
42.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.025 ± 1.657 | 2.319 ± 0.775 |
4.771 ± 0.403 | 7.753 ± 0.615 |
5.103 ± 0.938 | 5.5 ± 0.753 |
1.922 ± 0.382 | 3.777 ± 0.396 |
6.296 ± 1.191 | 10.404 ± 0.649 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.988 ± 0.285 | 5.036 ± 0.335 |
5.434 ± 0.898 | 4.705 ± 0.127 |
4.904 ± 0.318 | 5.898 ± 0.268 |
5.699 ± 0.653 | 6.494 ± 0.665 |
1.458 ± 0.429 | 3.512 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
