
Flaviflexus sp. H23T48
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Flaviflexus; unclassified Flaviflexus
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
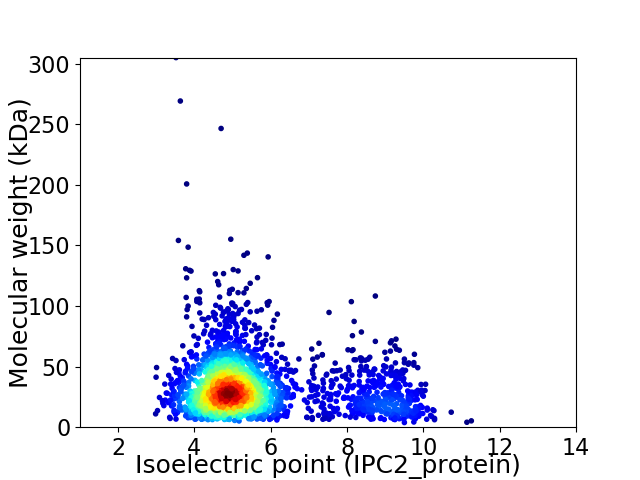
Virtual 2D-PAGE plot for 2334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
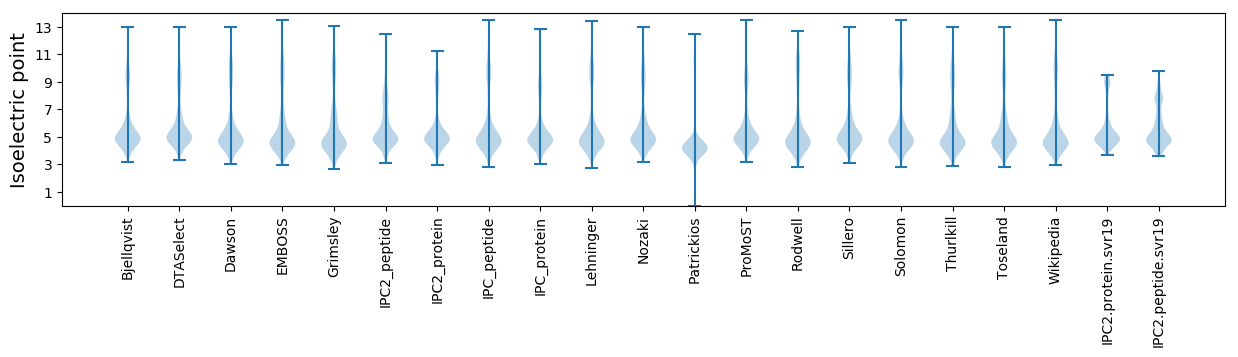
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9G680|A0A3Q9G680_9ACTO ABC transporter ATP-binding protein OS=Flaviflexus sp. H23T48 OX=2496867 GN=EJ997_01705 PE=4 SV=1
MM1 pKa = 7.26TSHH4 pKa = 6.47VPGSRR9 pKa = 11.84RR10 pKa = 11.84TLRR13 pKa = 11.84ACIATLGAGALGLGAFIVPAAATPSADD40 pKa = 4.13PIYY43 pKa = 10.6EE44 pKa = 4.15SDD46 pKa = 3.54PSVLVGSSANQASVAGSAYY65 pKa = 10.01GSRR68 pKa = 11.84ATGEE72 pKa = 4.08WFVSFSSAPLVNGGTQARR90 pKa = 11.84VDD92 pKa = 3.36ADD94 pKa = 3.99LNEE97 pKa = 4.52LEE99 pKa = 4.87SAIADD104 pKa = 3.57LDD106 pKa = 4.17VEE108 pKa = 5.0VKK110 pKa = 10.61ASHH113 pKa = 6.01SEE115 pKa = 3.69LWAGAVVTADD125 pKa = 3.6DD126 pKa = 4.24AEE128 pKa = 4.33LAKK131 pKa = 11.03VLDD134 pKa = 4.21RR135 pKa = 11.84VDD137 pKa = 3.39VAGVFPVLLIDD148 pKa = 4.75APEE151 pKa = 3.79QRR153 pKa = 11.84DD154 pKa = 3.61LGAEE158 pKa = 3.88PEE160 pKa = 4.08MRR162 pKa = 11.84FAAGMTGADD171 pKa = 3.75FAQDD175 pKa = 3.97LGVSGYY181 pKa = 10.18GQTISIIDD189 pKa = 3.46TGVDD193 pKa = 2.93IDD195 pKa = 4.0HH196 pKa = 6.49VAFGGTGVADD206 pKa = 3.62TTPWSEE212 pKa = 3.82INGNGSRR219 pKa = 11.84IIGGYY224 pKa = 10.58DD225 pKa = 3.71FVGDD229 pKa = 4.72DD230 pKa = 3.86YY231 pKa = 11.65NANPAADD238 pKa = 3.97NYY240 pKa = 10.91NPVPAPDD247 pKa = 4.75SNPDD251 pKa = 3.3DD252 pKa = 4.22CQGHH256 pKa = 5.16GTHH259 pKa = 6.15VAGIAAGSDD268 pKa = 3.64DD269 pKa = 3.63VTGVYY274 pKa = 10.32GVAPDD279 pKa = 4.24ANILAYY285 pKa = 10.02RR286 pKa = 11.84VFGCEE291 pKa = 3.62GSTDD295 pKa = 3.66SAIMLQAMEE304 pKa = 4.23ASARR308 pKa = 11.84DD309 pKa = 3.35GADD312 pKa = 2.78IVNMSIGAGFMTWPNYY328 pKa = 5.66PTAVAADD335 pKa = 3.82NLVDD339 pKa = 3.76AGVVVTVSQGNEE351 pKa = 3.9GEE353 pKa = 4.37SGVFSGGAPAVASKK367 pKa = 10.4VISVGSVDD375 pKa = 3.76NVASEE380 pKa = 4.43APVFVVNDD388 pKa = 3.23QNIAYY393 pKa = 9.52SSSTGAPNPPTDD405 pKa = 3.32NSEE408 pKa = 4.29YY409 pKa = 10.85EE410 pKa = 4.3IIAYY414 pKa = 7.91PAGSEE419 pKa = 4.01TGAVPIEE426 pKa = 4.23DD427 pKa = 4.1AEE429 pKa = 4.49GKK431 pKa = 9.85VVIVRR436 pKa = 11.84RR437 pKa = 11.84GNSTFHH443 pKa = 6.6EE444 pKa = 4.49KK445 pKa = 9.97AAAAQEE451 pKa = 4.21SGAAGVIIDD460 pKa = 4.35NNVAGAINATVEE472 pKa = 4.05GDD474 pKa = 3.58PAITIPTVTISMADD488 pKa = 3.21GDD490 pKa = 5.16AIRR493 pKa = 11.84ADD495 pKa = 3.43LAAAEE500 pKa = 4.43EE501 pKa = 4.76PVTLTWSDD509 pKa = 3.31QTEE512 pKa = 4.1VSEE515 pKa = 4.25IATGGLVSDD524 pKa = 4.72FSSYY528 pKa = 11.09GLSADD533 pKa = 3.82LTVKK537 pKa = 9.97PQVVAPGGFINSAYY551 pKa = 9.16PLEE554 pKa = 4.9AGPSGYY560 pKa = 8.93ATLSGTSMAAPHH572 pKa = 5.79VAGAAALILEE582 pKa = 4.83VNPALNPSQVLTSLQNTADD601 pKa = 3.86PLLWSGNPDD610 pKa = 3.2LGFPEE615 pKa = 4.48PVHH618 pKa = 6.07RR619 pKa = 11.84QGAGMINIPNAIFANLWGTQVTPSQINLYY648 pKa = 10.96DD649 pKa = 3.83NDD651 pKa = 3.88TGASEE656 pKa = 4.21THH658 pKa = 6.3TLTIHH663 pKa = 6.26NRR665 pKa = 11.84DD666 pKa = 3.44EE667 pKa = 4.56VEE669 pKa = 4.06FTYY672 pKa = 11.1SFVNLAGIATAGINQLPNYY691 pKa = 8.87YY692 pKa = 9.94GAAATTEE699 pKa = 4.66FSAEE703 pKa = 4.25TVTVPAGGTATVDD716 pKa = 3.36VTITEE721 pKa = 4.34PEE723 pKa = 4.25GFAGGIYY730 pKa = 10.16GGQIVVLGEE739 pKa = 4.08DD740 pKa = 3.12ASGYY744 pKa = 4.54TTQSIVSYY752 pKa = 10.67GGLAGDD758 pKa = 4.59YY759 pKa = 7.32EE760 pKa = 4.44TDD762 pKa = 3.52LAFMEE767 pKa = 4.02QWTYY771 pKa = 11.14RR772 pKa = 11.84DD773 pKa = 3.22YY774 pKa = 11.41GYY776 pKa = 9.13TEE778 pKa = 4.29EE779 pKa = 4.81EE780 pKa = 3.97VGEE783 pKa = 4.84LIDD786 pKa = 3.82EE787 pKa = 5.62PIYY790 pKa = 9.68STPGLGVLEE799 pKa = 5.16DD800 pKa = 3.75CTKK803 pKa = 10.83LVDD806 pKa = 4.09GEE808 pKa = 4.64CVSEE812 pKa = 5.13GNYY815 pKa = 10.29SWGHH819 pKa = 5.93DD820 pKa = 3.34EE821 pKa = 4.44YY822 pKa = 11.27VYY824 pKa = 11.52SMEE827 pKa = 6.48DD828 pKa = 3.09GDD830 pKa = 3.8TPYY833 pKa = 11.39AVLHH837 pKa = 6.36IEE839 pKa = 4.25NPVSYY844 pKa = 9.78MKK846 pKa = 10.41IEE848 pKa = 4.28AFHH851 pKa = 6.9ANADD855 pKa = 3.86GTKK858 pKa = 9.77GDD860 pKa = 4.54PVSPDD865 pKa = 3.14YY866 pKa = 11.38NVVYY870 pKa = 10.57EE871 pKa = 4.29SDD873 pKa = 3.87GEE875 pKa = 4.45GAVPGIQVYY884 pKa = 9.22QWDD887 pKa = 3.88GTYY890 pKa = 10.76VKK892 pKa = 10.58SADD895 pKa = 3.34SEE897 pKa = 4.61QYY899 pKa = 11.06LKK901 pKa = 11.06ALDD904 pKa = 3.79GDD906 pKa = 4.31YY907 pKa = 10.83VIEE910 pKa = 4.18VTAVKK915 pKa = 10.67GLGSIDD921 pKa = 3.9NPDD924 pKa = 3.59HH925 pKa = 7.3VEE927 pKa = 3.88VWTSPAFTIDD937 pKa = 3.33RR938 pKa = 11.84DD939 pKa = 4.11GVPGEE944 pKa = 4.32GPGDD948 pKa = 3.64GSIDD952 pKa = 3.54EE953 pKa = 4.53EE954 pKa = 4.24ADD956 pKa = 3.4KK957 pKa = 11.61GFFLYY962 pKa = 11.01NDD964 pKa = 3.92FEE966 pKa = 4.73SAADD970 pKa = 3.51HH971 pKa = 5.94YY972 pKa = 11.47FMYY975 pKa = 10.54GRR977 pKa = 11.84DD978 pKa = 3.52GDD980 pKa = 4.19EE981 pKa = 4.29VFVGDD986 pKa = 4.12WDD988 pKa = 4.13ADD990 pKa = 3.84GVDD993 pKa = 3.94TIGVRR998 pKa = 11.84RR999 pKa = 11.84GHH1001 pKa = 5.62TFYY1004 pKa = 11.25VQNQLIGGNAEE1015 pKa = 3.88VEE1017 pKa = 4.81FKK1019 pKa = 10.73FGRR1022 pKa = 11.84DD1023 pKa = 2.91GDD1025 pKa = 4.34EE1026 pKa = 4.09ILVGDD1031 pKa = 4.83WNADD1035 pKa = 3.32GYY1037 pKa = 10.48DD1038 pKa = 3.39TFAVRR1043 pKa = 11.84RR1044 pKa = 11.84GVTIYY1049 pKa = 10.7AQNEE1053 pKa = 4.58LIGGNAEE1060 pKa = 3.95VEE1062 pKa = 4.56FKK1064 pKa = 10.65YY1065 pKa = 11.07GRR1067 pKa = 11.84VGDD1070 pKa = 4.54DD1071 pKa = 4.11LFAGDD1076 pKa = 4.53WDD1078 pKa = 3.85GDD1080 pKa = 4.09GYY1082 pKa = 10.67DD1083 pKa = 3.35TFAVRR1088 pKa = 11.84RR1089 pKa = 11.84GNSFYY1094 pKa = 11.13VQNSLVGGNAEE1105 pKa = 4.05DD1106 pKa = 3.9TFHH1109 pKa = 7.28FGHH1112 pKa = 6.72EE1113 pKa = 4.34SDD1115 pKa = 3.81TVLVGDD1121 pKa = 4.15WDD1123 pKa = 4.29GDD1125 pKa = 4.01LKK1127 pKa = 11.08DD1128 pKa = 3.74TVALRR1133 pKa = 11.84RR1134 pKa = 11.84GDD1136 pKa = 3.42TFYY1139 pKa = 11.56VSNSLSSGSADD1150 pKa = 3.03RR1151 pKa = 11.84SFVLGRR1157 pKa = 11.84DD1158 pKa = 3.18SDD1160 pKa = 4.07EE1161 pKa = 4.35VLVGDD1166 pKa = 4.31WNGDD1170 pKa = 3.76GIDD1173 pKa = 3.68SVGLRR1178 pKa = 11.84RR1179 pKa = 4.23
MM1 pKa = 7.26TSHH4 pKa = 6.47VPGSRR9 pKa = 11.84RR10 pKa = 11.84TLRR13 pKa = 11.84ACIATLGAGALGLGAFIVPAAATPSADD40 pKa = 4.13PIYY43 pKa = 10.6EE44 pKa = 4.15SDD46 pKa = 3.54PSVLVGSSANQASVAGSAYY65 pKa = 10.01GSRR68 pKa = 11.84ATGEE72 pKa = 4.08WFVSFSSAPLVNGGTQARR90 pKa = 11.84VDD92 pKa = 3.36ADD94 pKa = 3.99LNEE97 pKa = 4.52LEE99 pKa = 4.87SAIADD104 pKa = 3.57LDD106 pKa = 4.17VEE108 pKa = 5.0VKK110 pKa = 10.61ASHH113 pKa = 6.01SEE115 pKa = 3.69LWAGAVVTADD125 pKa = 3.6DD126 pKa = 4.24AEE128 pKa = 4.33LAKK131 pKa = 11.03VLDD134 pKa = 4.21RR135 pKa = 11.84VDD137 pKa = 3.39VAGVFPVLLIDD148 pKa = 4.75APEE151 pKa = 3.79QRR153 pKa = 11.84DD154 pKa = 3.61LGAEE158 pKa = 3.88PEE160 pKa = 4.08MRR162 pKa = 11.84FAAGMTGADD171 pKa = 3.75FAQDD175 pKa = 3.97LGVSGYY181 pKa = 10.18GQTISIIDD189 pKa = 3.46TGVDD193 pKa = 2.93IDD195 pKa = 4.0HH196 pKa = 6.49VAFGGTGVADD206 pKa = 3.62TTPWSEE212 pKa = 3.82INGNGSRR219 pKa = 11.84IIGGYY224 pKa = 10.58DD225 pKa = 3.71FVGDD229 pKa = 4.72DD230 pKa = 3.86YY231 pKa = 11.65NANPAADD238 pKa = 3.97NYY240 pKa = 10.91NPVPAPDD247 pKa = 4.75SNPDD251 pKa = 3.3DD252 pKa = 4.22CQGHH256 pKa = 5.16GTHH259 pKa = 6.15VAGIAAGSDD268 pKa = 3.64DD269 pKa = 3.63VTGVYY274 pKa = 10.32GVAPDD279 pKa = 4.24ANILAYY285 pKa = 10.02RR286 pKa = 11.84VFGCEE291 pKa = 3.62GSTDD295 pKa = 3.66SAIMLQAMEE304 pKa = 4.23ASARR308 pKa = 11.84DD309 pKa = 3.35GADD312 pKa = 2.78IVNMSIGAGFMTWPNYY328 pKa = 5.66PTAVAADD335 pKa = 3.82NLVDD339 pKa = 3.76AGVVVTVSQGNEE351 pKa = 3.9GEE353 pKa = 4.37SGVFSGGAPAVASKK367 pKa = 10.4VISVGSVDD375 pKa = 3.76NVASEE380 pKa = 4.43APVFVVNDD388 pKa = 3.23QNIAYY393 pKa = 9.52SSSTGAPNPPTDD405 pKa = 3.32NSEE408 pKa = 4.29YY409 pKa = 10.85EE410 pKa = 4.3IIAYY414 pKa = 7.91PAGSEE419 pKa = 4.01TGAVPIEE426 pKa = 4.23DD427 pKa = 4.1AEE429 pKa = 4.49GKK431 pKa = 9.85VVIVRR436 pKa = 11.84RR437 pKa = 11.84GNSTFHH443 pKa = 6.6EE444 pKa = 4.49KK445 pKa = 9.97AAAAQEE451 pKa = 4.21SGAAGVIIDD460 pKa = 4.35NNVAGAINATVEE472 pKa = 4.05GDD474 pKa = 3.58PAITIPTVTISMADD488 pKa = 3.21GDD490 pKa = 5.16AIRR493 pKa = 11.84ADD495 pKa = 3.43LAAAEE500 pKa = 4.43EE501 pKa = 4.76PVTLTWSDD509 pKa = 3.31QTEE512 pKa = 4.1VSEE515 pKa = 4.25IATGGLVSDD524 pKa = 4.72FSSYY528 pKa = 11.09GLSADD533 pKa = 3.82LTVKK537 pKa = 9.97PQVVAPGGFINSAYY551 pKa = 9.16PLEE554 pKa = 4.9AGPSGYY560 pKa = 8.93ATLSGTSMAAPHH572 pKa = 5.79VAGAAALILEE582 pKa = 4.83VNPALNPSQVLTSLQNTADD601 pKa = 3.86PLLWSGNPDD610 pKa = 3.2LGFPEE615 pKa = 4.48PVHH618 pKa = 6.07RR619 pKa = 11.84QGAGMINIPNAIFANLWGTQVTPSQINLYY648 pKa = 10.96DD649 pKa = 3.83NDD651 pKa = 3.88TGASEE656 pKa = 4.21THH658 pKa = 6.3TLTIHH663 pKa = 6.26NRR665 pKa = 11.84DD666 pKa = 3.44EE667 pKa = 4.56VEE669 pKa = 4.06FTYY672 pKa = 11.1SFVNLAGIATAGINQLPNYY691 pKa = 8.87YY692 pKa = 9.94GAAATTEE699 pKa = 4.66FSAEE703 pKa = 4.25TVTVPAGGTATVDD716 pKa = 3.36VTITEE721 pKa = 4.34PEE723 pKa = 4.25GFAGGIYY730 pKa = 10.16GGQIVVLGEE739 pKa = 4.08DD740 pKa = 3.12ASGYY744 pKa = 4.54TTQSIVSYY752 pKa = 10.67GGLAGDD758 pKa = 4.59YY759 pKa = 7.32EE760 pKa = 4.44TDD762 pKa = 3.52LAFMEE767 pKa = 4.02QWTYY771 pKa = 11.14RR772 pKa = 11.84DD773 pKa = 3.22YY774 pKa = 11.41GYY776 pKa = 9.13TEE778 pKa = 4.29EE779 pKa = 4.81EE780 pKa = 3.97VGEE783 pKa = 4.84LIDD786 pKa = 3.82EE787 pKa = 5.62PIYY790 pKa = 9.68STPGLGVLEE799 pKa = 5.16DD800 pKa = 3.75CTKK803 pKa = 10.83LVDD806 pKa = 4.09GEE808 pKa = 4.64CVSEE812 pKa = 5.13GNYY815 pKa = 10.29SWGHH819 pKa = 5.93DD820 pKa = 3.34EE821 pKa = 4.44YY822 pKa = 11.27VYY824 pKa = 11.52SMEE827 pKa = 6.48DD828 pKa = 3.09GDD830 pKa = 3.8TPYY833 pKa = 11.39AVLHH837 pKa = 6.36IEE839 pKa = 4.25NPVSYY844 pKa = 9.78MKK846 pKa = 10.41IEE848 pKa = 4.28AFHH851 pKa = 6.9ANADD855 pKa = 3.86GTKK858 pKa = 9.77GDD860 pKa = 4.54PVSPDD865 pKa = 3.14YY866 pKa = 11.38NVVYY870 pKa = 10.57EE871 pKa = 4.29SDD873 pKa = 3.87GEE875 pKa = 4.45GAVPGIQVYY884 pKa = 9.22QWDD887 pKa = 3.88GTYY890 pKa = 10.76VKK892 pKa = 10.58SADD895 pKa = 3.34SEE897 pKa = 4.61QYY899 pKa = 11.06LKK901 pKa = 11.06ALDD904 pKa = 3.79GDD906 pKa = 4.31YY907 pKa = 10.83VIEE910 pKa = 4.18VTAVKK915 pKa = 10.67GLGSIDD921 pKa = 3.9NPDD924 pKa = 3.59HH925 pKa = 7.3VEE927 pKa = 3.88VWTSPAFTIDD937 pKa = 3.33RR938 pKa = 11.84DD939 pKa = 4.11GVPGEE944 pKa = 4.32GPGDD948 pKa = 3.64GSIDD952 pKa = 3.54EE953 pKa = 4.53EE954 pKa = 4.24ADD956 pKa = 3.4KK957 pKa = 11.61GFFLYY962 pKa = 11.01NDD964 pKa = 3.92FEE966 pKa = 4.73SAADD970 pKa = 3.51HH971 pKa = 5.94YY972 pKa = 11.47FMYY975 pKa = 10.54GRR977 pKa = 11.84DD978 pKa = 3.52GDD980 pKa = 4.19EE981 pKa = 4.29VFVGDD986 pKa = 4.12WDD988 pKa = 4.13ADD990 pKa = 3.84GVDD993 pKa = 3.94TIGVRR998 pKa = 11.84RR999 pKa = 11.84GHH1001 pKa = 5.62TFYY1004 pKa = 11.25VQNQLIGGNAEE1015 pKa = 3.88VEE1017 pKa = 4.81FKK1019 pKa = 10.73FGRR1022 pKa = 11.84DD1023 pKa = 2.91GDD1025 pKa = 4.34EE1026 pKa = 4.09ILVGDD1031 pKa = 4.83WNADD1035 pKa = 3.32GYY1037 pKa = 10.48DD1038 pKa = 3.39TFAVRR1043 pKa = 11.84RR1044 pKa = 11.84GVTIYY1049 pKa = 10.7AQNEE1053 pKa = 4.58LIGGNAEE1060 pKa = 3.95VEE1062 pKa = 4.56FKK1064 pKa = 10.65YY1065 pKa = 11.07GRR1067 pKa = 11.84VGDD1070 pKa = 4.54DD1071 pKa = 4.11LFAGDD1076 pKa = 4.53WDD1078 pKa = 3.85GDD1080 pKa = 4.09GYY1082 pKa = 10.67DD1083 pKa = 3.35TFAVRR1088 pKa = 11.84RR1089 pKa = 11.84GNSFYY1094 pKa = 11.13VQNSLVGGNAEE1105 pKa = 4.05DD1106 pKa = 3.9TFHH1109 pKa = 7.28FGHH1112 pKa = 6.72EE1113 pKa = 4.34SDD1115 pKa = 3.81TVLVGDD1121 pKa = 4.15WDD1123 pKa = 4.29GDD1125 pKa = 4.01LKK1127 pKa = 11.08DD1128 pKa = 3.74TVALRR1133 pKa = 11.84RR1134 pKa = 11.84GDD1136 pKa = 3.42TFYY1139 pKa = 11.56VSNSLSSGSADD1150 pKa = 3.03RR1151 pKa = 11.84SFVLGRR1157 pKa = 11.84DD1158 pKa = 3.18SDD1160 pKa = 4.07EE1161 pKa = 4.35VLVGDD1166 pKa = 4.31WNGDD1170 pKa = 3.76GIDD1173 pKa = 3.68SVGLRR1178 pKa = 11.84RR1179 pKa = 4.23
Molecular weight: 123.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9G536|A0A3Q9G536_9ACTO Riboflavin synthase OS=Flaviflexus sp. H23T48 OX=2496867 GN=EJ997_09760 PE=4 SV=1
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.78RR13 pKa = 11.84AKK15 pKa = 9.18VHH17 pKa = 5.49GFRR20 pKa = 11.84KK21 pKa = 9.95RR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILSGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.9GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 8.78RR13 pKa = 11.84AKK15 pKa = 9.18VHH17 pKa = 5.49GFRR20 pKa = 11.84KK21 pKa = 9.95RR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILSGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.9GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
747752 |
32 |
2932 |
320.4 |
34.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.771 ± 0.054 | 0.647 ± 0.015 |
6.36 ± 0.047 | 6.808 ± 0.05 |
3.135 ± 0.031 | 8.593 ± 0.05 |
1.964 ± 0.022 | 5.619 ± 0.036 |
3.143 ± 0.038 | 9.424 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.241 ± 0.024 | 2.853 ± 0.026 |
4.989 ± 0.034 | 2.976 ± 0.027 |
6.03 ± 0.053 | 6.282 ± 0.039 |
6.26 ± 0.048 | 8.132 ± 0.047 |
1.434 ± 0.02 | 2.34 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
