
Human adenovirus D serotype 17 (HAdV-17) (Human adenovirus 17)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus; Human mastadenovirus D
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
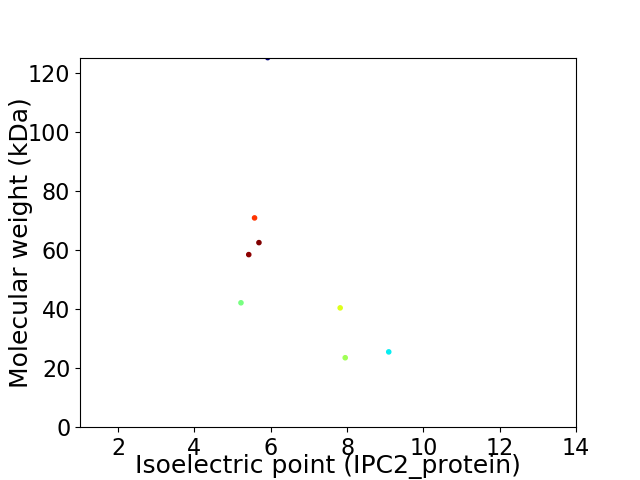
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9WF16|Q9WF16_ADE17 Pre-hexon-linking protein IIIa OS=Human adenovirus D serotype 17 OX=46922 PE=2 SV=1
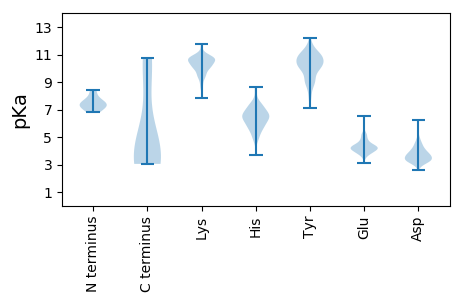
MM1 pKa = 7.31HH2 pKa = 7.36PVLRR6 pKa = 11.84QMRR9 pKa = 11.84PTPPATTATAAVAGAGASQPQPQTEE34 pKa = 3.87MDD36 pKa = 3.68LEE38 pKa = 4.42EE39 pKa = 6.32GEE41 pKa = 4.35GLARR45 pKa = 11.84LGAPSPEE52 pKa = 3.6RR53 pKa = 11.84HH54 pKa = 6.33PRR56 pKa = 11.84VQLQKK61 pKa = 10.54DD62 pKa = 3.51VRR64 pKa = 11.84PAYY67 pKa = 10.69VPAQNLFRR75 pKa = 11.84DD76 pKa = 3.85RR77 pKa = 11.84SGEE80 pKa = 3.95EE81 pKa = 3.87PEE83 pKa = 4.09EE84 pKa = 3.87MRR86 pKa = 11.84DD87 pKa = 3.31CRR89 pKa = 11.84FRR91 pKa = 11.84AGRR94 pKa = 11.84EE95 pKa = 3.76LRR97 pKa = 11.84EE98 pKa = 3.94GLDD101 pKa = 3.31RR102 pKa = 11.84QRR104 pKa = 11.84VLRR107 pKa = 11.84DD108 pKa = 2.89EE109 pKa = 5.21DD110 pKa = 4.09FEE112 pKa = 5.17PNEE115 pKa = 3.9QTGISPARR123 pKa = 11.84AHH125 pKa = 5.74VAAANLVTAYY135 pKa = 9.0EE136 pKa = 4.04QTVKK140 pKa = 10.49QEE142 pKa = 4.4RR143 pKa = 11.84NFQKK147 pKa = 10.59SFNNHH152 pKa = 3.71VRR154 pKa = 11.84TLIARR159 pKa = 11.84EE160 pKa = 4.02EE161 pKa = 4.21VALGLMHH168 pKa = 7.41LWDD171 pKa = 3.96LAEE174 pKa = 5.92AIVQNPDD181 pKa = 3.41SKK183 pKa = 11.15PLTAQLFLVVQHH195 pKa = 6.48SRR197 pKa = 11.84DD198 pKa = 3.36NEE200 pKa = 3.99AFRR203 pKa = 11.84EE204 pKa = 4.03ALLNIAEE211 pKa = 4.33PEE213 pKa = 4.24GRR215 pKa = 11.84WLLEE219 pKa = 4.86LINILQSIVVQEE231 pKa = 4.36RR232 pKa = 11.84SLSLAEE238 pKa = 4.47KK239 pKa = 10.11VAAINYY245 pKa = 9.0SVLSLGKK252 pKa = 9.39FYY254 pKa = 11.12ARR256 pKa = 11.84KK257 pKa = 9.39IYY259 pKa = 7.63KK260 pKa = 8.73TPYY263 pKa = 8.86VPIDD267 pKa = 3.65KK268 pKa = 9.91EE269 pKa = 4.28VKK271 pKa = 9.4IDD273 pKa = 3.6SFYY276 pKa = 10.94MRR278 pKa = 11.84MALKK282 pKa = 10.67VLTLSDD288 pKa = 3.71DD289 pKa = 3.79LGVYY293 pKa = 10.56RR294 pKa = 11.84NDD296 pKa = 4.41RR297 pKa = 11.84IHH299 pKa = 6.67KK300 pKa = 9.47AVSASRR306 pKa = 11.84RR307 pKa = 11.84RR308 pKa = 11.84EE309 pKa = 3.75LSDD312 pKa = 3.68RR313 pKa = 11.84EE314 pKa = 4.15LMLSLRR320 pKa = 11.84RR321 pKa = 11.84ALVGGAAGGEE331 pKa = 3.78EE332 pKa = 5.21SYY334 pKa = 11.63FDD336 pKa = 4.05MGADD340 pKa = 3.61LHH342 pKa = 5.83WQPSRR347 pKa = 11.84RR348 pKa = 11.84ALEE351 pKa = 3.94AAYY354 pKa = 10.02GPEE357 pKa = 4.18DD358 pKa = 3.77LDD360 pKa = 3.74EE361 pKa = 4.65EE362 pKa = 4.49EE363 pKa = 5.14EE364 pKa = 4.29EE365 pKa = 5.12EE366 pKa = 4.28EE367 pKa = 4.16DD368 pKa = 4.86APAAGYY374 pKa = 10.02
MM1 pKa = 7.31HH2 pKa = 7.36PVLRR6 pKa = 11.84QMRR9 pKa = 11.84PTPPATTATAAVAGAGASQPQPQTEE34 pKa = 3.87MDD36 pKa = 3.68LEE38 pKa = 4.42EE39 pKa = 6.32GEE41 pKa = 4.35GLARR45 pKa = 11.84LGAPSPEE52 pKa = 3.6RR53 pKa = 11.84HH54 pKa = 6.33PRR56 pKa = 11.84VQLQKK61 pKa = 10.54DD62 pKa = 3.51VRR64 pKa = 11.84PAYY67 pKa = 10.69VPAQNLFRR75 pKa = 11.84DD76 pKa = 3.85RR77 pKa = 11.84SGEE80 pKa = 3.95EE81 pKa = 3.87PEE83 pKa = 4.09EE84 pKa = 3.87MRR86 pKa = 11.84DD87 pKa = 3.31CRR89 pKa = 11.84FRR91 pKa = 11.84AGRR94 pKa = 11.84EE95 pKa = 3.76LRR97 pKa = 11.84EE98 pKa = 3.94GLDD101 pKa = 3.31RR102 pKa = 11.84QRR104 pKa = 11.84VLRR107 pKa = 11.84DD108 pKa = 2.89EE109 pKa = 5.21DD110 pKa = 4.09FEE112 pKa = 5.17PNEE115 pKa = 3.9QTGISPARR123 pKa = 11.84AHH125 pKa = 5.74VAAANLVTAYY135 pKa = 9.0EE136 pKa = 4.04QTVKK140 pKa = 10.49QEE142 pKa = 4.4RR143 pKa = 11.84NFQKK147 pKa = 10.59SFNNHH152 pKa = 3.71VRR154 pKa = 11.84TLIARR159 pKa = 11.84EE160 pKa = 4.02EE161 pKa = 4.21VALGLMHH168 pKa = 7.41LWDD171 pKa = 3.96LAEE174 pKa = 5.92AIVQNPDD181 pKa = 3.41SKK183 pKa = 11.15PLTAQLFLVVQHH195 pKa = 6.48SRR197 pKa = 11.84DD198 pKa = 3.36NEE200 pKa = 3.99AFRR203 pKa = 11.84EE204 pKa = 4.03ALLNIAEE211 pKa = 4.33PEE213 pKa = 4.24GRR215 pKa = 11.84WLLEE219 pKa = 4.86LINILQSIVVQEE231 pKa = 4.36RR232 pKa = 11.84SLSLAEE238 pKa = 4.47KK239 pKa = 10.11VAAINYY245 pKa = 9.0SVLSLGKK252 pKa = 9.39FYY254 pKa = 11.12ARR256 pKa = 11.84KK257 pKa = 9.39IYY259 pKa = 7.63KK260 pKa = 8.73TPYY263 pKa = 8.86VPIDD267 pKa = 3.65KK268 pKa = 9.91EE269 pKa = 4.28VKK271 pKa = 9.4IDD273 pKa = 3.6SFYY276 pKa = 10.94MRR278 pKa = 11.84MALKK282 pKa = 10.67VLTLSDD288 pKa = 3.71DD289 pKa = 3.79LGVYY293 pKa = 10.56RR294 pKa = 11.84NDD296 pKa = 4.41RR297 pKa = 11.84IHH299 pKa = 6.67KK300 pKa = 9.47AVSASRR306 pKa = 11.84RR307 pKa = 11.84RR308 pKa = 11.84EE309 pKa = 3.75LSDD312 pKa = 3.68RR313 pKa = 11.84EE314 pKa = 4.15LMLSLRR320 pKa = 11.84RR321 pKa = 11.84ALVGGAAGGEE331 pKa = 3.78EE332 pKa = 5.21SYY334 pKa = 11.63FDD336 pKa = 4.05MGADD340 pKa = 3.61LHH342 pKa = 5.83WQPSRR347 pKa = 11.84RR348 pKa = 11.84ALEE351 pKa = 3.94AAYY354 pKa = 10.02GPEE357 pKa = 4.18DD358 pKa = 3.77LDD360 pKa = 3.74EE361 pKa = 4.65EE362 pKa = 4.49EE363 pKa = 5.14EE364 pKa = 4.29EE365 pKa = 5.12EE366 pKa = 4.28EE367 pKa = 4.16DD368 pKa = 4.86APAAGYY374 pKa = 10.02
Molecular weight: 42.19 kDa
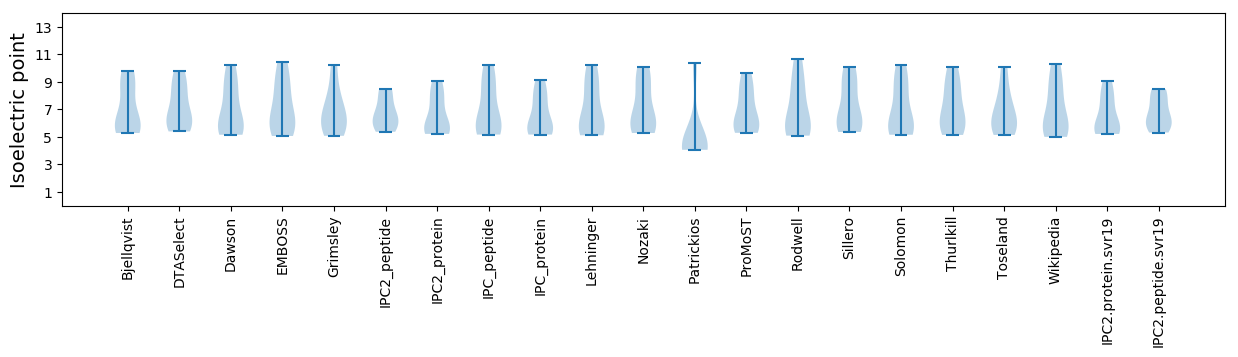
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9WF18|Q9WF18_ADE17 Isoform of Q9WF19 Pre-protein VI OS=Human adenovirus D serotype 17 OX=46922 GN=L3 PE=2 SV=1
MM1 pKa = 7.8EE2 pKa = 6.49DD3 pKa = 3.25INFASLAPRR12 pKa = 11.84HH13 pKa = 5.03GTRR16 pKa = 11.84PFMGTWNEE24 pKa = 3.89IGTSQLNGGAFNWSSVWSGLKK45 pKa = 10.58NFGSTLRR52 pKa = 11.84TYY54 pKa = 11.16GNKK57 pKa = 9.37AWNSSTGQLLRR68 pKa = 11.84EE69 pKa = 4.1KK70 pKa = 10.86LKK72 pKa = 10.96DD73 pKa = 3.29QNFQQKK79 pKa = 9.05VVDD82 pKa = 4.39GLASGINGVVDD93 pKa = 3.77IANQAVQRR101 pKa = 11.84EE102 pKa = 4.58INSRR106 pKa = 11.84LDD108 pKa = 3.31PRR110 pKa = 11.84PPTVVEE116 pKa = 4.43MEE118 pKa = 4.73DD119 pKa = 3.33ATLPPPKK126 pKa = 10.05GEE128 pKa = 3.9KK129 pKa = 9.84RR130 pKa = 11.84PRR132 pKa = 11.84PDD134 pKa = 3.14AEE136 pKa = 4.04EE137 pKa = 4.59TILQVDD143 pKa = 4.62EE144 pKa = 4.54PPSYY148 pKa = 11.1EE149 pKa = 4.14EE150 pKa = 3.88AVKK153 pKa = 10.86AGMPTTRR160 pKa = 11.84IIAPLATGVMKK171 pKa = 10.09PATLDD176 pKa = 3.54LPPPPTPAPPKK187 pKa = 9.6AAPVVQPPPVATAVRR202 pKa = 11.84RR203 pKa = 11.84VPARR207 pKa = 11.84RR208 pKa = 11.84QAQNWQSTLHH218 pKa = 6.39SIVGLGVKK226 pKa = 8.75SLKK229 pKa = 9.96RR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84CYY234 pKa = 10.71
MM1 pKa = 7.8EE2 pKa = 6.49DD3 pKa = 3.25INFASLAPRR12 pKa = 11.84HH13 pKa = 5.03GTRR16 pKa = 11.84PFMGTWNEE24 pKa = 3.89IGTSQLNGGAFNWSSVWSGLKK45 pKa = 10.58NFGSTLRR52 pKa = 11.84TYY54 pKa = 11.16GNKK57 pKa = 9.37AWNSSTGQLLRR68 pKa = 11.84EE69 pKa = 4.1KK70 pKa = 10.86LKK72 pKa = 10.96DD73 pKa = 3.29QNFQQKK79 pKa = 9.05VVDD82 pKa = 4.39GLASGINGVVDD93 pKa = 3.77IANQAVQRR101 pKa = 11.84EE102 pKa = 4.58INSRR106 pKa = 11.84LDD108 pKa = 3.31PRR110 pKa = 11.84PPTVVEE116 pKa = 4.43MEE118 pKa = 4.73DD119 pKa = 3.33ATLPPPKK126 pKa = 10.05GEE128 pKa = 3.9KK129 pKa = 9.84RR130 pKa = 11.84PRR132 pKa = 11.84PDD134 pKa = 3.14AEE136 pKa = 4.04EE137 pKa = 4.59TILQVDD143 pKa = 4.62EE144 pKa = 4.54PPSYY148 pKa = 11.1EE149 pKa = 4.14EE150 pKa = 3.88AVKK153 pKa = 10.86AGMPTTRR160 pKa = 11.84IIAPLATGVMKK171 pKa = 10.09PATLDD176 pKa = 3.54LPPPPTPAPPKK187 pKa = 9.6AAPVVQPPPVATAVRR202 pKa = 11.84RR203 pKa = 11.84VPARR207 pKa = 11.84RR208 pKa = 11.84QAQNWQSTLHH218 pKa = 6.39SIVGLGVKK226 pKa = 8.75SLKK229 pKa = 9.96RR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84CYY234 pKa = 10.71
Molecular weight: 25.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3971 |
209 |
1091 |
496.4 |
56.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.26 ± 0.645 | 1.158 ± 0.36 |
5.767 ± 0.228 | 6.673 ± 0.679 |
4.155 ± 0.366 | 5.54 ± 0.447 |
1.989 ± 0.381 | 3.526 ± 0.219 |
3.651 ± 0.861 | 9.872 ± 0.364 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.217 | 4.382 ± 0.517 |
6.799 ± 0.378 | 4.231 ± 0.512 |
7.958 ± 0.942 | 6.422 ± 0.554 |
5.465 ± 0.517 | 6.724 ± 0.388 |
1.461 ± 0.187 | 3.576 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
