
Desulfobacula toluolica (strain DSM 7467 / Tol2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfobacula; Desulfobacula toluolica
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
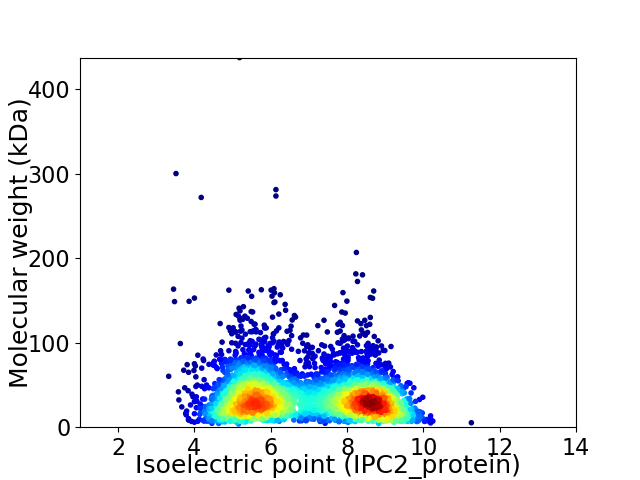
Virtual 2D-PAGE plot for 4189 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0NHQ9|K0NHQ9_DESTT Putative membrane protein OS=Desulfobacula toluolica (strain DSM 7467 / Tol2) OX=651182 GN=TOL2_C03370 PE=3 SV=1
MM1 pKa = 7.68AIDD4 pKa = 4.02SNIITDD10 pKa = 4.03MGQLSALTYY19 pKa = 10.53KK20 pKa = 10.6NYY22 pKa = 10.03PSEE25 pKa = 4.33SSNSLLNKK33 pKa = 10.23GKK35 pKa = 10.1ILQGTFNIGQDD46 pKa = 3.37SFSLNNSYY54 pKa = 7.62TVKK57 pKa = 10.61DD58 pKa = 3.98YY59 pKa = 11.75ADD61 pKa = 4.12TPSDD65 pKa = 3.6MQALLLEE72 pKa = 5.1KK73 pKa = 10.38NDD75 pKa = 3.56STGNPTGEE83 pKa = 3.98YY84 pKa = 9.6IIAFRR89 pKa = 11.84GTQEE93 pKa = 3.96KK94 pKa = 10.04MDD96 pKa = 3.94IGVDD100 pKa = 4.17AIIGLANYY108 pKa = 9.42NPQFNDD114 pKa = 2.5AKK116 pKa = 11.15AFVQQMMTDD125 pKa = 3.84HH126 pKa = 6.92NISSSNLTLTGHH138 pKa = 6.01SLGAILTQSVGAVLGIKK155 pKa = 10.2GYY157 pKa = 10.95AYY159 pKa = 10.64NPYY162 pKa = 8.69GTEE165 pKa = 3.78RR166 pKa = 11.84LLTMWEE172 pKa = 4.57SYY174 pKa = 10.21TDD176 pKa = 3.69SLGEE180 pKa = 3.83ALIQVGIYY188 pKa = 9.68QVLNAFGLDD197 pKa = 3.1SSYY200 pKa = 11.92AQFAADD206 pKa = 3.84NILNVSFNDD215 pKa = 3.6CGTLNGGILSNFASEE230 pKa = 4.31LTSDD234 pKa = 3.74HH235 pKa = 6.87LGTYY239 pKa = 10.47LPVFGDD245 pKa = 3.65NEE247 pKa = 4.33GLSGHH252 pKa = 5.99SMVVLNNAISYY263 pKa = 8.1YY264 pKa = 10.73NEE266 pKa = 4.3IIAHH270 pKa = 5.59FTDD273 pKa = 3.27EE274 pKa = 4.19TDD276 pKa = 3.68YY277 pKa = 11.99DD278 pKa = 4.26DD279 pKa = 6.58LSTAYY284 pKa = 10.15ALSGEE289 pKa = 4.23NGFNRR294 pKa = 11.84LNNTFGKK301 pKa = 10.56LDD303 pKa = 3.39IAHH306 pKa = 7.19AAGNSLQFKK315 pKa = 10.13FLDD318 pKa = 3.79KK319 pKa = 11.3SSITDD324 pKa = 3.79FQSQASDD331 pKa = 3.47QAHH334 pKa = 6.61LFSLRR339 pKa = 11.84ALNPFAIIGANYY351 pKa = 10.28SIVNEE356 pKa = 4.1NGEE359 pKa = 4.42LDD361 pKa = 3.25IDD363 pKa = 4.07NYY365 pKa = 9.3SDD367 pKa = 4.47KK368 pKa = 11.15YY369 pKa = 11.0IEE371 pKa = 4.63DD372 pKa = 3.56RR373 pKa = 11.84STFLYY378 pKa = 10.82YY379 pKa = 10.57LAHH382 pKa = 7.34PDD384 pKa = 3.21EE385 pKa = 5.06KK386 pKa = 10.98LPSGEE391 pKa = 4.25DD392 pKa = 3.32TIQFTDD398 pKa = 3.06KK399 pKa = 10.8RR400 pKa = 11.84LGINTTAWKK409 pKa = 10.46SGIGVDD415 pKa = 3.99LTIRR419 pKa = 11.84DD420 pKa = 4.34YY421 pKa = 11.36FWGTEE426 pKa = 3.72VRR428 pKa = 11.84DD429 pKa = 3.76EE430 pKa = 4.52FGSNGNTGDD439 pKa = 3.5DD440 pKa = 3.72HH441 pKa = 8.14LYY443 pKa = 11.39GMGGNDD449 pKa = 3.32TLKK452 pKa = 10.93GYY454 pKa = 10.57GGEE457 pKa = 4.34DD458 pKa = 3.81YY459 pKa = 11.06IEE461 pKa = 4.67GGEE464 pKa = 4.55GQDD467 pKa = 3.14TMYY470 pKa = 11.25GGGDD474 pKa = 3.25KK475 pKa = 10.38DD476 pKa = 3.35TFYY479 pKa = 10.84IQGEE483 pKa = 4.09DD484 pKa = 3.44DD485 pKa = 5.58DD486 pKa = 5.86YY487 pKa = 11.75DD488 pKa = 3.6IFNGGDD494 pKa = 3.48HH495 pKa = 6.96NEE497 pKa = 3.92DD498 pKa = 3.91TILGSEE504 pKa = 4.35GNDD507 pKa = 3.58TIRR510 pKa = 11.84VHH512 pKa = 7.1DD513 pKa = 4.22FSGEE517 pKa = 4.0NTVEE521 pKa = 4.31IIDD524 pKa = 3.74GRR526 pKa = 11.84GGEE529 pKa = 4.08NVIAGTGMVDD539 pKa = 3.9TIDD542 pKa = 3.44MSGTALIDD550 pKa = 3.08IDD552 pKa = 4.84RR553 pKa = 11.84IEE555 pKa = 5.03GGDD558 pKa = 3.62GADD561 pKa = 3.73TIKK564 pKa = 11.13GCLADD569 pKa = 3.61DD570 pKa = 4.87TIYY573 pKa = 11.07GGSKK577 pKa = 10.42DD578 pKa = 3.86QIEE581 pKa = 4.78DD582 pKa = 3.49NAVDD586 pKa = 3.79QLEE589 pKa = 4.58GGAGNDD595 pKa = 2.77IYY597 pKa = 11.51YY598 pKa = 10.54AGTGDD603 pKa = 5.01VINDD607 pKa = 3.45LDD609 pKa = 3.94GRR611 pKa = 11.84GTVWFEE617 pKa = 3.44GRR619 pKa = 11.84EE620 pKa = 4.04LSGLTWTGLSPDD632 pKa = 3.54SNIYY636 pKa = 10.29SDD638 pKa = 4.19SDD640 pKa = 3.35EE641 pKa = 4.53NYY643 pKa = 10.43YY644 pKa = 11.44ALFDD648 pKa = 3.79NTSNTLIVSHH658 pKa = 6.96SSTNHH663 pKa = 5.52YY664 pKa = 10.44IKK666 pKa = 10.32IEE668 pKa = 4.01NFSDD672 pKa = 3.5GTLGLTLEE680 pKa = 5.44DD681 pKa = 3.81YY682 pKa = 8.91TPPSDD687 pKa = 4.25YY688 pKa = 10.82DD689 pKa = 3.46YY690 pKa = 11.1TLIGSADD697 pKa = 3.92DD698 pKa = 5.26DD699 pKa = 4.31EE700 pKa = 7.07SDD702 pKa = 3.54YY703 pKa = 11.65HH704 pKa = 7.14PDD706 pKa = 3.51YY707 pKa = 11.72DD708 pKa = 4.92LGTYY712 pKa = 10.12SYY714 pKa = 11.59GFGDD718 pKa = 3.91NADD721 pKa = 3.27ITADD725 pKa = 3.34MSTSISLEE733 pKa = 3.85IYY735 pKa = 10.18GGAGSDD741 pKa = 4.17YY742 pKa = 10.96ILGLPWDD749 pKa = 4.93DD750 pKa = 3.78YY751 pKa = 11.95LNGGGGDD758 pKa = 3.63DD759 pKa = 4.88HH760 pKa = 7.98IVSNSNSTMAPDD772 pKa = 3.3IVGDD776 pKa = 3.9VMDD779 pKa = 5.36GGDD782 pKa = 4.35GNDD785 pKa = 4.9LIQDD789 pKa = 4.02TGNVGSVMLGGAGFDD804 pKa = 3.15ILNGYY809 pKa = 8.96RR810 pKa = 11.84GNDD813 pKa = 3.32TMSGGSQTDD822 pKa = 3.49VLTGHH827 pKa = 7.26AGDD830 pKa = 5.47DD831 pKa = 3.89YY832 pKa = 11.78LSGGDD837 pKa = 4.01GNDD840 pKa = 3.22VLLGDD845 pKa = 5.34NDD847 pKa = 4.67LFWTNSIMGMLGPNMVDD864 pKa = 3.72FTFDD868 pKa = 3.07QAGWITDD875 pKa = 3.51VNFNGAMSVKK885 pKa = 10.5DD886 pKa = 3.47GDD888 pKa = 4.44TITVTGIGGGDD899 pKa = 3.17IYY901 pKa = 11.32FDD903 pKa = 3.48IPAGNDD909 pKa = 3.28FLDD912 pKa = 4.14GGNGNDD918 pKa = 4.02HH919 pKa = 7.1LYY921 pKa = 10.94DD922 pKa = 3.4
MM1 pKa = 7.68AIDD4 pKa = 4.02SNIITDD10 pKa = 4.03MGQLSALTYY19 pKa = 10.53KK20 pKa = 10.6NYY22 pKa = 10.03PSEE25 pKa = 4.33SSNSLLNKK33 pKa = 10.23GKK35 pKa = 10.1ILQGTFNIGQDD46 pKa = 3.37SFSLNNSYY54 pKa = 7.62TVKK57 pKa = 10.61DD58 pKa = 3.98YY59 pKa = 11.75ADD61 pKa = 4.12TPSDD65 pKa = 3.6MQALLLEE72 pKa = 5.1KK73 pKa = 10.38NDD75 pKa = 3.56STGNPTGEE83 pKa = 3.98YY84 pKa = 9.6IIAFRR89 pKa = 11.84GTQEE93 pKa = 3.96KK94 pKa = 10.04MDD96 pKa = 3.94IGVDD100 pKa = 4.17AIIGLANYY108 pKa = 9.42NPQFNDD114 pKa = 2.5AKK116 pKa = 11.15AFVQQMMTDD125 pKa = 3.84HH126 pKa = 6.92NISSSNLTLTGHH138 pKa = 6.01SLGAILTQSVGAVLGIKK155 pKa = 10.2GYY157 pKa = 10.95AYY159 pKa = 10.64NPYY162 pKa = 8.69GTEE165 pKa = 3.78RR166 pKa = 11.84LLTMWEE172 pKa = 4.57SYY174 pKa = 10.21TDD176 pKa = 3.69SLGEE180 pKa = 3.83ALIQVGIYY188 pKa = 9.68QVLNAFGLDD197 pKa = 3.1SSYY200 pKa = 11.92AQFAADD206 pKa = 3.84NILNVSFNDD215 pKa = 3.6CGTLNGGILSNFASEE230 pKa = 4.31LTSDD234 pKa = 3.74HH235 pKa = 6.87LGTYY239 pKa = 10.47LPVFGDD245 pKa = 3.65NEE247 pKa = 4.33GLSGHH252 pKa = 5.99SMVVLNNAISYY263 pKa = 8.1YY264 pKa = 10.73NEE266 pKa = 4.3IIAHH270 pKa = 5.59FTDD273 pKa = 3.27EE274 pKa = 4.19TDD276 pKa = 3.68YY277 pKa = 11.99DD278 pKa = 4.26DD279 pKa = 6.58LSTAYY284 pKa = 10.15ALSGEE289 pKa = 4.23NGFNRR294 pKa = 11.84LNNTFGKK301 pKa = 10.56LDD303 pKa = 3.39IAHH306 pKa = 7.19AAGNSLQFKK315 pKa = 10.13FLDD318 pKa = 3.79KK319 pKa = 11.3SSITDD324 pKa = 3.79FQSQASDD331 pKa = 3.47QAHH334 pKa = 6.61LFSLRR339 pKa = 11.84ALNPFAIIGANYY351 pKa = 10.28SIVNEE356 pKa = 4.1NGEE359 pKa = 4.42LDD361 pKa = 3.25IDD363 pKa = 4.07NYY365 pKa = 9.3SDD367 pKa = 4.47KK368 pKa = 11.15YY369 pKa = 11.0IEE371 pKa = 4.63DD372 pKa = 3.56RR373 pKa = 11.84STFLYY378 pKa = 10.82YY379 pKa = 10.57LAHH382 pKa = 7.34PDD384 pKa = 3.21EE385 pKa = 5.06KK386 pKa = 10.98LPSGEE391 pKa = 4.25DD392 pKa = 3.32TIQFTDD398 pKa = 3.06KK399 pKa = 10.8RR400 pKa = 11.84LGINTTAWKK409 pKa = 10.46SGIGVDD415 pKa = 3.99LTIRR419 pKa = 11.84DD420 pKa = 4.34YY421 pKa = 11.36FWGTEE426 pKa = 3.72VRR428 pKa = 11.84DD429 pKa = 3.76EE430 pKa = 4.52FGSNGNTGDD439 pKa = 3.5DD440 pKa = 3.72HH441 pKa = 8.14LYY443 pKa = 11.39GMGGNDD449 pKa = 3.32TLKK452 pKa = 10.93GYY454 pKa = 10.57GGEE457 pKa = 4.34DD458 pKa = 3.81YY459 pKa = 11.06IEE461 pKa = 4.67GGEE464 pKa = 4.55GQDD467 pKa = 3.14TMYY470 pKa = 11.25GGGDD474 pKa = 3.25KK475 pKa = 10.38DD476 pKa = 3.35TFYY479 pKa = 10.84IQGEE483 pKa = 4.09DD484 pKa = 3.44DD485 pKa = 5.58DD486 pKa = 5.86YY487 pKa = 11.75DD488 pKa = 3.6IFNGGDD494 pKa = 3.48HH495 pKa = 6.96NEE497 pKa = 3.92DD498 pKa = 3.91TILGSEE504 pKa = 4.35GNDD507 pKa = 3.58TIRR510 pKa = 11.84VHH512 pKa = 7.1DD513 pKa = 4.22FSGEE517 pKa = 4.0NTVEE521 pKa = 4.31IIDD524 pKa = 3.74GRR526 pKa = 11.84GGEE529 pKa = 4.08NVIAGTGMVDD539 pKa = 3.9TIDD542 pKa = 3.44MSGTALIDD550 pKa = 3.08IDD552 pKa = 4.84RR553 pKa = 11.84IEE555 pKa = 5.03GGDD558 pKa = 3.62GADD561 pKa = 3.73TIKK564 pKa = 11.13GCLADD569 pKa = 3.61DD570 pKa = 4.87TIYY573 pKa = 11.07GGSKK577 pKa = 10.42DD578 pKa = 3.86QIEE581 pKa = 4.78DD582 pKa = 3.49NAVDD586 pKa = 3.79QLEE589 pKa = 4.58GGAGNDD595 pKa = 2.77IYY597 pKa = 11.51YY598 pKa = 10.54AGTGDD603 pKa = 5.01VINDD607 pKa = 3.45LDD609 pKa = 3.94GRR611 pKa = 11.84GTVWFEE617 pKa = 3.44GRR619 pKa = 11.84EE620 pKa = 4.04LSGLTWTGLSPDD632 pKa = 3.54SNIYY636 pKa = 10.29SDD638 pKa = 4.19SDD640 pKa = 3.35EE641 pKa = 4.53NYY643 pKa = 10.43YY644 pKa = 11.44ALFDD648 pKa = 3.79NTSNTLIVSHH658 pKa = 6.96SSTNHH663 pKa = 5.52YY664 pKa = 10.44IKK666 pKa = 10.32IEE668 pKa = 4.01NFSDD672 pKa = 3.5GTLGLTLEE680 pKa = 5.44DD681 pKa = 3.81YY682 pKa = 8.91TPPSDD687 pKa = 4.25YY688 pKa = 10.82DD689 pKa = 3.46YY690 pKa = 11.1TLIGSADD697 pKa = 3.92DD698 pKa = 5.26DD699 pKa = 4.31EE700 pKa = 7.07SDD702 pKa = 3.54YY703 pKa = 11.65HH704 pKa = 7.14PDD706 pKa = 3.51YY707 pKa = 11.72DD708 pKa = 4.92LGTYY712 pKa = 10.12SYY714 pKa = 11.59GFGDD718 pKa = 3.91NADD721 pKa = 3.27ITADD725 pKa = 3.34MSTSISLEE733 pKa = 3.85IYY735 pKa = 10.18GGAGSDD741 pKa = 4.17YY742 pKa = 10.96ILGLPWDD749 pKa = 4.93DD750 pKa = 3.78YY751 pKa = 11.95LNGGGGDD758 pKa = 3.63DD759 pKa = 4.88HH760 pKa = 7.98IVSNSNSTMAPDD772 pKa = 3.3IVGDD776 pKa = 3.9VMDD779 pKa = 5.36GGDD782 pKa = 4.35GNDD785 pKa = 4.9LIQDD789 pKa = 4.02TGNVGSVMLGGAGFDD804 pKa = 3.15ILNGYY809 pKa = 8.96RR810 pKa = 11.84GNDD813 pKa = 3.32TMSGGSQTDD822 pKa = 3.49VLTGHH827 pKa = 7.26AGDD830 pKa = 5.47DD831 pKa = 3.89YY832 pKa = 11.78LSGGDD837 pKa = 4.01GNDD840 pKa = 3.22VLLGDD845 pKa = 5.34NDD847 pKa = 4.67LFWTNSIMGMLGPNMVDD864 pKa = 3.72FTFDD868 pKa = 3.07QAGWITDD875 pKa = 3.51VNFNGAMSVKK885 pKa = 10.5DD886 pKa = 3.47GDD888 pKa = 4.44TITVTGIGGGDD899 pKa = 3.17IYY901 pKa = 11.32FDD903 pKa = 3.48IPAGNDD909 pKa = 3.28FLDD912 pKa = 4.14GGNGNDD918 pKa = 4.02HH919 pKa = 7.1LYY921 pKa = 10.94DD922 pKa = 3.4
Molecular weight: 99.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0NQ50|K0NQ50_DESTT BamI2: uncharacterized anaerobic dehydrogenase 2Fe-2S and 4Fe-4S ferredoxin iron-sulfur protein OS=Desulfobacula toluolica (strain DSM 7467 / Tol2) OX=651182 GN=bamI2 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 7.64RR12 pKa = 11.84TRR14 pKa = 11.84KK15 pKa = 9.43HH16 pKa = 4.96GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTPSGRR28 pKa = 11.84RR29 pKa = 11.84VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.86GRR39 pKa = 11.84KK40 pKa = 8.94KK41 pKa = 10.21LTVV44 pKa = 3.1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 7.64RR12 pKa = 11.84TRR14 pKa = 11.84KK15 pKa = 9.43HH16 pKa = 4.96GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTPSGRR28 pKa = 11.84RR29 pKa = 11.84VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.86GRR39 pKa = 11.84KK40 pKa = 8.94KK41 pKa = 10.21LTVV44 pKa = 3.1
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1459019 |
32 |
3944 |
348.3 |
39.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.927 ± 0.04 | 1.381 ± 0.017 |
5.765 ± 0.031 | 6.2 ± 0.036 |
4.827 ± 0.028 | 6.696 ± 0.058 |
2.046 ± 0.015 | 8.156 ± 0.036 |
7.804 ± 0.043 | 9.475 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.756 ± 0.016 | 4.645 ± 0.026 |
3.849 ± 0.023 | 3.489 ± 0.02 |
4.265 ± 0.025 | 6.227 ± 0.028 |
5.249 ± 0.025 | 6.165 ± 0.026 |
0.96 ± 0.014 | 3.118 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
