
Salipiger mucosus DSM 16094
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Salipiger; Salipiger mucosus
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
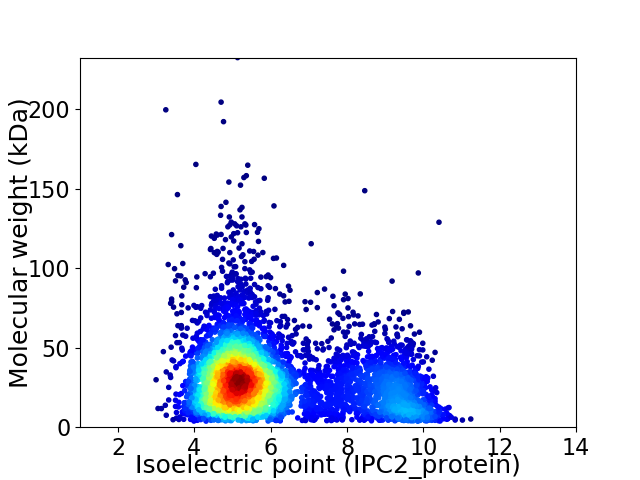
Virtual 2D-PAGE plot for 5583 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S9R544|S9R544_9RHOB Peptide deformylase OS=Salipiger mucosus DSM 16094 OX=1123237 GN=def PE=3 SV=1
MM1 pKa = 7.86PEE3 pKa = 3.5AFLRR7 pKa = 11.84GVANLVRR14 pKa = 11.84GFDD17 pKa = 3.63RR18 pKa = 11.84LVSVAMSTHH27 pKa = 6.89PNGQANTQKK36 pKa = 10.75GEE38 pKa = 4.3SYY40 pKa = 9.37MATYY44 pKa = 10.22YY45 pKa = 11.17VSTTGSNANDD55 pKa = 3.87GSEE58 pKa = 4.08SSPFATINHH67 pKa = 6.26AVSSGLQPGDD77 pKa = 3.37TVLVKK82 pKa = 10.39PGTYY86 pKa = 9.3RR87 pKa = 11.84EE88 pKa = 4.09SVYY91 pKa = 11.35VNIDD95 pKa = 2.96GSAAGDD101 pKa = 3.15ITIRR105 pKa = 11.84SEE107 pKa = 4.29TPGAASIQPGSSGNGFTIEE126 pKa = 4.06GSYY129 pKa = 8.82ITIDD133 pKa = 3.23GFEE136 pKa = 4.21ITGSAYY142 pKa = 10.35HH143 pKa = 6.99GIEE146 pKa = 4.23AQNSHH151 pKa = 7.18HH152 pKa = 6.98ISMLNNISHH161 pKa = 7.62DD162 pKa = 3.79NGASGLSAIWGEE174 pKa = 4.18FYY176 pKa = 10.78LIEE179 pKa = 4.98GNTTYY184 pKa = 11.45GNAASDD190 pKa = 3.8WFSGISVYY198 pKa = 8.77EE199 pKa = 3.87NRR201 pKa = 11.84NITGDD206 pKa = 3.72TEE208 pKa = 4.33TTGYY212 pKa = 8.48RR213 pKa = 11.84TIIRR217 pKa = 11.84NNVSYY222 pKa = 11.51DD223 pKa = 3.2NVTEE227 pKa = 4.35NGQHH231 pKa = 5.92TDD233 pKa = 3.01GNGIIIDD240 pKa = 4.52DD241 pKa = 4.14FQSTQNSAFPSYY253 pKa = 10.25TYY255 pKa = 7.92PTLVEE260 pKa = 4.07NNLVYY265 pKa = 10.91EE266 pKa = 4.3NGGKK270 pKa = 10.14GIQVIWSDD278 pKa = 3.24NVTVRR283 pKa = 11.84GNTAYY288 pKa = 10.66HH289 pKa = 6.38NNQDD293 pKa = 3.83LQNTGTWRR301 pKa = 11.84GEE303 pKa = 3.68ISNSQSSNNTFVNNIAVVDD322 pKa = 4.15PSVHH326 pKa = 6.62PEE328 pKa = 3.36NTAIDD333 pKa = 3.72NNSYY337 pKa = 11.07GGYY340 pKa = 9.05TNDD343 pKa = 3.05NVVWANNLTYY353 pKa = 10.45TGTAGEE359 pKa = 4.59ASVKK363 pKa = 9.89TDD365 pKa = 3.61GGNAMPSAADD375 pKa = 3.72GNLLGVDD382 pKa = 4.38PGFIDD387 pKa = 3.7PGNDD391 pKa = 2.82FRR393 pKa = 11.84LSSGSVAIDD402 pKa = 3.93AGTDD406 pKa = 3.48DD407 pKa = 4.45YY408 pKa = 12.1GLASVDD414 pKa = 3.68LEE416 pKa = 4.04GTARR420 pKa = 11.84VEE422 pKa = 4.17GVVDD426 pKa = 3.6IGAYY430 pKa = 9.79EE431 pKa = 4.33RR432 pKa = 11.84DD433 pKa = 3.6SSGTPTPPANTAPEE447 pKa = 4.12ATNDD451 pKa = 3.51SGFSTTEE458 pKa = 3.55DD459 pKa = 3.19TAATIATADD468 pKa = 3.72LRR470 pKa = 11.84ANDD473 pKa = 3.86SDD475 pKa = 4.51ADD477 pKa = 3.86GDD479 pKa = 4.27ALTVTSVSGAVNGTVSLASASSVVFTPDD507 pKa = 2.62AGYY510 pKa = 10.99SGMASFDD517 pKa = 3.64YY518 pKa = 10.32TVSDD522 pKa = 4.06GAGGTDD528 pKa = 3.15TATVSIEE535 pKa = 3.84VTAASAPPSNTAPVATDD552 pKa = 3.32DD553 pKa = 4.26SGFATSEE560 pKa = 3.92DD561 pKa = 3.73TALSLKK567 pKa = 10.6PSMLKK572 pKa = 10.98DD573 pKa = 3.27NDD575 pKa = 3.28TDD577 pKa = 4.17ADD579 pKa = 4.21GDD581 pKa = 4.24SLTITNVSGAVNGTVSLAADD601 pKa = 3.78EE602 pKa = 4.77TVVFTPDD609 pKa = 2.81AGFTGMARR617 pKa = 11.84FDD619 pKa = 3.66YY620 pKa = 10.14TISDD624 pKa = 4.01GAGGTDD630 pKa = 3.2TAQAEE635 pKa = 4.63IQVSTAEE642 pKa = 4.22VTPPAPPEE650 pKa = 4.3DD651 pKa = 4.31DD652 pKa = 4.14EE653 pKa = 4.61EE654 pKa = 6.07AGSFSIWDD662 pKa = 3.51SSATPDD668 pKa = 4.27IMADD672 pKa = 3.19SDD674 pKa = 3.99TAAVTLGLRR683 pKa = 11.84FTADD687 pKa = 2.29VDD689 pKa = 3.88AALEE693 pKa = 3.88ALRR696 pKa = 11.84LYY698 pKa = 10.8VSEE701 pKa = 5.28ANTGTQSINLWSDD714 pKa = 2.48SGTRR718 pKa = 11.84LGQATAEE725 pKa = 4.27VTEE728 pKa = 4.9SGWSTVSFDD737 pKa = 5.48KK738 pKa = 10.8PLQLEE743 pKa = 4.01AGEE746 pKa = 5.17DD747 pKa = 4.24YY748 pKa = 10.57IASYY752 pKa = 9.49YY753 pKa = 10.15APNGGYY759 pKa = 9.52VVSEE763 pKa = 5.08DD764 pKa = 4.11YY765 pKa = 10.82FDD767 pKa = 4.0SASDD771 pKa = 3.44EE772 pKa = 4.48GPISLEE778 pKa = 4.16ANSGVYY784 pKa = 10.1SYY786 pKa = 11.82GSGGSSFPDD795 pKa = 2.97QSYY798 pKa = 10.8AFSNYY803 pKa = 7.35WVDD806 pKa = 4.23VVLTPDD812 pKa = 3.19TATDD816 pKa = 3.54VAGDD820 pKa = 3.78NVIGEE825 pKa = 4.54TGTVVAGQAGAGDD838 pKa = 3.92WTSVTFDD845 pKa = 4.1TPLEE849 pKa = 4.5DD850 pKa = 3.8PSVVMGGLTGNGANPYY866 pKa = 6.71TVRR869 pKa = 11.84VRR871 pKa = 11.84NVTDD875 pKa = 3.03TGFEE879 pKa = 4.08YY880 pKa = 10.72QIDD883 pKa = 4.15EE884 pKa = 4.6YY885 pKa = 11.24EE886 pKa = 4.54YY887 pKa = 11.23LDD889 pKa = 3.95DD890 pKa = 3.31WHH892 pKa = 6.0TTEE895 pKa = 4.8EE896 pKa = 4.29ISWLAVEE903 pKa = 5.02KK904 pKa = 9.54GTHH907 pKa = 4.97VLSDD911 pKa = 3.33GRR913 pKa = 11.84TISAGDD919 pKa = 3.6ASVGGSFAGISFDD932 pKa = 5.67DD933 pKa = 4.5GAFDD937 pKa = 3.62SAPVVLGQAVGEE949 pKa = 4.16ANAMAVTDD957 pKa = 3.98RR958 pKa = 11.84LRR960 pKa = 11.84SVSEE964 pKa = 3.9TGFEE968 pKa = 3.84ARR970 pKa = 11.84LFQQEE975 pKa = 4.09AATGSIEE982 pKa = 4.98AEE984 pKa = 4.07GFDD987 pKa = 4.59WIAVGQGGSATDD999 pKa = 3.6GALAGTSGNMVDD1011 pKa = 4.56HH1012 pKa = 7.22RR1013 pKa = 11.84PTTVDD1018 pKa = 3.63FDD1020 pKa = 4.46GSFSGEE1026 pKa = 4.21DD1027 pKa = 3.41FVMVTDD1033 pKa = 3.96MQTTNGWDD1041 pKa = 3.24VATLQTASLDD1051 pKa = 3.86DD1052 pKa = 3.91EE1053 pKa = 5.58SMVLRR1058 pKa = 11.84VLEE1061 pKa = 4.09EE1062 pKa = 3.7TSRR1065 pKa = 11.84DD1066 pKa = 3.39GEE1068 pKa = 4.3VRR1070 pKa = 11.84HH1071 pKa = 5.91VDD1073 pKa = 3.06EE1074 pKa = 5.41DD1075 pKa = 3.69AGFVGFEE1082 pKa = 3.93TGLIEE1087 pKa = 4.17GTPFEE1092 pKa = 4.89SDD1094 pKa = 3.03TVLL1097 pKa = 4.05
MM1 pKa = 7.86PEE3 pKa = 3.5AFLRR7 pKa = 11.84GVANLVRR14 pKa = 11.84GFDD17 pKa = 3.63RR18 pKa = 11.84LVSVAMSTHH27 pKa = 6.89PNGQANTQKK36 pKa = 10.75GEE38 pKa = 4.3SYY40 pKa = 9.37MATYY44 pKa = 10.22YY45 pKa = 11.17VSTTGSNANDD55 pKa = 3.87GSEE58 pKa = 4.08SSPFATINHH67 pKa = 6.26AVSSGLQPGDD77 pKa = 3.37TVLVKK82 pKa = 10.39PGTYY86 pKa = 9.3RR87 pKa = 11.84EE88 pKa = 4.09SVYY91 pKa = 11.35VNIDD95 pKa = 2.96GSAAGDD101 pKa = 3.15ITIRR105 pKa = 11.84SEE107 pKa = 4.29TPGAASIQPGSSGNGFTIEE126 pKa = 4.06GSYY129 pKa = 8.82ITIDD133 pKa = 3.23GFEE136 pKa = 4.21ITGSAYY142 pKa = 10.35HH143 pKa = 6.99GIEE146 pKa = 4.23AQNSHH151 pKa = 7.18HH152 pKa = 6.98ISMLNNISHH161 pKa = 7.62DD162 pKa = 3.79NGASGLSAIWGEE174 pKa = 4.18FYY176 pKa = 10.78LIEE179 pKa = 4.98GNTTYY184 pKa = 11.45GNAASDD190 pKa = 3.8WFSGISVYY198 pKa = 8.77EE199 pKa = 3.87NRR201 pKa = 11.84NITGDD206 pKa = 3.72TEE208 pKa = 4.33TTGYY212 pKa = 8.48RR213 pKa = 11.84TIIRR217 pKa = 11.84NNVSYY222 pKa = 11.51DD223 pKa = 3.2NVTEE227 pKa = 4.35NGQHH231 pKa = 5.92TDD233 pKa = 3.01GNGIIIDD240 pKa = 4.52DD241 pKa = 4.14FQSTQNSAFPSYY253 pKa = 10.25TYY255 pKa = 7.92PTLVEE260 pKa = 4.07NNLVYY265 pKa = 10.91EE266 pKa = 4.3NGGKK270 pKa = 10.14GIQVIWSDD278 pKa = 3.24NVTVRR283 pKa = 11.84GNTAYY288 pKa = 10.66HH289 pKa = 6.38NNQDD293 pKa = 3.83LQNTGTWRR301 pKa = 11.84GEE303 pKa = 3.68ISNSQSSNNTFVNNIAVVDD322 pKa = 4.15PSVHH326 pKa = 6.62PEE328 pKa = 3.36NTAIDD333 pKa = 3.72NNSYY337 pKa = 11.07GGYY340 pKa = 9.05TNDD343 pKa = 3.05NVVWANNLTYY353 pKa = 10.45TGTAGEE359 pKa = 4.59ASVKK363 pKa = 9.89TDD365 pKa = 3.61GGNAMPSAADD375 pKa = 3.72GNLLGVDD382 pKa = 4.38PGFIDD387 pKa = 3.7PGNDD391 pKa = 2.82FRR393 pKa = 11.84LSSGSVAIDD402 pKa = 3.93AGTDD406 pKa = 3.48DD407 pKa = 4.45YY408 pKa = 12.1GLASVDD414 pKa = 3.68LEE416 pKa = 4.04GTARR420 pKa = 11.84VEE422 pKa = 4.17GVVDD426 pKa = 3.6IGAYY430 pKa = 9.79EE431 pKa = 4.33RR432 pKa = 11.84DD433 pKa = 3.6SSGTPTPPANTAPEE447 pKa = 4.12ATNDD451 pKa = 3.51SGFSTTEE458 pKa = 3.55DD459 pKa = 3.19TAATIATADD468 pKa = 3.72LRR470 pKa = 11.84ANDD473 pKa = 3.86SDD475 pKa = 4.51ADD477 pKa = 3.86GDD479 pKa = 4.27ALTVTSVSGAVNGTVSLASASSVVFTPDD507 pKa = 2.62AGYY510 pKa = 10.99SGMASFDD517 pKa = 3.64YY518 pKa = 10.32TVSDD522 pKa = 4.06GAGGTDD528 pKa = 3.15TATVSIEE535 pKa = 3.84VTAASAPPSNTAPVATDD552 pKa = 3.32DD553 pKa = 4.26SGFATSEE560 pKa = 3.92DD561 pKa = 3.73TALSLKK567 pKa = 10.6PSMLKK572 pKa = 10.98DD573 pKa = 3.27NDD575 pKa = 3.28TDD577 pKa = 4.17ADD579 pKa = 4.21GDD581 pKa = 4.24SLTITNVSGAVNGTVSLAADD601 pKa = 3.78EE602 pKa = 4.77TVVFTPDD609 pKa = 2.81AGFTGMARR617 pKa = 11.84FDD619 pKa = 3.66YY620 pKa = 10.14TISDD624 pKa = 4.01GAGGTDD630 pKa = 3.2TAQAEE635 pKa = 4.63IQVSTAEE642 pKa = 4.22VTPPAPPEE650 pKa = 4.3DD651 pKa = 4.31DD652 pKa = 4.14EE653 pKa = 4.61EE654 pKa = 6.07AGSFSIWDD662 pKa = 3.51SSATPDD668 pKa = 4.27IMADD672 pKa = 3.19SDD674 pKa = 3.99TAAVTLGLRR683 pKa = 11.84FTADD687 pKa = 2.29VDD689 pKa = 3.88AALEE693 pKa = 3.88ALRR696 pKa = 11.84LYY698 pKa = 10.8VSEE701 pKa = 5.28ANTGTQSINLWSDD714 pKa = 2.48SGTRR718 pKa = 11.84LGQATAEE725 pKa = 4.27VTEE728 pKa = 4.9SGWSTVSFDD737 pKa = 5.48KK738 pKa = 10.8PLQLEE743 pKa = 4.01AGEE746 pKa = 5.17DD747 pKa = 4.24YY748 pKa = 10.57IASYY752 pKa = 9.49YY753 pKa = 10.15APNGGYY759 pKa = 9.52VVSEE763 pKa = 5.08DD764 pKa = 4.11YY765 pKa = 10.82FDD767 pKa = 4.0SASDD771 pKa = 3.44EE772 pKa = 4.48GPISLEE778 pKa = 4.16ANSGVYY784 pKa = 10.1SYY786 pKa = 11.82GSGGSSFPDD795 pKa = 2.97QSYY798 pKa = 10.8AFSNYY803 pKa = 7.35WVDD806 pKa = 4.23VVLTPDD812 pKa = 3.19TATDD816 pKa = 3.54VAGDD820 pKa = 3.78NVIGEE825 pKa = 4.54TGTVVAGQAGAGDD838 pKa = 3.92WTSVTFDD845 pKa = 4.1TPLEE849 pKa = 4.5DD850 pKa = 3.8PSVVMGGLTGNGANPYY866 pKa = 6.71TVRR869 pKa = 11.84VRR871 pKa = 11.84NVTDD875 pKa = 3.03TGFEE879 pKa = 4.08YY880 pKa = 10.72QIDD883 pKa = 4.15EE884 pKa = 4.6YY885 pKa = 11.24EE886 pKa = 4.54YY887 pKa = 11.23LDD889 pKa = 3.95DD890 pKa = 3.31WHH892 pKa = 6.0TTEE895 pKa = 4.8EE896 pKa = 4.29ISWLAVEE903 pKa = 5.02KK904 pKa = 9.54GTHH907 pKa = 4.97VLSDD911 pKa = 3.33GRR913 pKa = 11.84TISAGDD919 pKa = 3.6ASVGGSFAGISFDD932 pKa = 5.67DD933 pKa = 4.5GAFDD937 pKa = 3.62SAPVVLGQAVGEE949 pKa = 4.16ANAMAVTDD957 pKa = 3.98RR958 pKa = 11.84LRR960 pKa = 11.84SVSEE964 pKa = 3.9TGFEE968 pKa = 3.84ARR970 pKa = 11.84LFQQEE975 pKa = 4.09AATGSIEE982 pKa = 4.98AEE984 pKa = 4.07GFDD987 pKa = 4.59WIAVGQGGSATDD999 pKa = 3.6GALAGTSGNMVDD1011 pKa = 4.56HH1012 pKa = 7.22RR1013 pKa = 11.84PTTVDD1018 pKa = 3.63FDD1020 pKa = 4.46GSFSGEE1026 pKa = 4.21DD1027 pKa = 3.41FVMVTDD1033 pKa = 3.96MQTTNGWDD1041 pKa = 3.24VATLQTASLDD1051 pKa = 3.86DD1052 pKa = 3.91EE1053 pKa = 5.58SMVLRR1058 pKa = 11.84VLEE1061 pKa = 4.09EE1062 pKa = 3.7TSRR1065 pKa = 11.84DD1066 pKa = 3.39GEE1068 pKa = 4.3VRR1070 pKa = 11.84HH1071 pKa = 5.91VDD1073 pKa = 3.06EE1074 pKa = 5.41DD1075 pKa = 3.69AGFVGFEE1082 pKa = 3.93TGLIEE1087 pKa = 4.17GTPFEE1092 pKa = 4.89SDD1094 pKa = 3.03TVLL1097 pKa = 4.05
Molecular weight: 114.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S9Q315|S9Q315_9RHOB L-fuconolactone hydrolase OS=Salipiger mucosus DSM 16094 OX=1123237 GN=Salmuc_01176 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44AGRR28 pKa = 11.84KK29 pKa = 8.46VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.79NLSAA44 pKa = 4.73
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44AGRR28 pKa = 11.84KK29 pKa = 8.46VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.79NLSAA44 pKa = 4.73
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1630836 |
37 |
2149 |
292.1 |
31.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.472 ± 0.049 | 0.893 ± 0.011 |
6.174 ± 0.033 | 6.521 ± 0.029 |
3.564 ± 0.022 | 8.975 ± 0.038 |
2.091 ± 0.017 | 4.642 ± 0.028 |
2.614 ± 0.029 | 9.909 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.741 ± 0.018 | 2.294 ± 0.02 |
5.295 ± 0.025 | 3.003 ± 0.02 |
7.493 ± 0.041 | 5.08 ± 0.026 |
5.399 ± 0.024 | 7.295 ± 0.029 |
1.402 ± 0.015 | 2.143 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
