
Wuhan insect virus 21
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.99
Get precalculated fractions of proteins
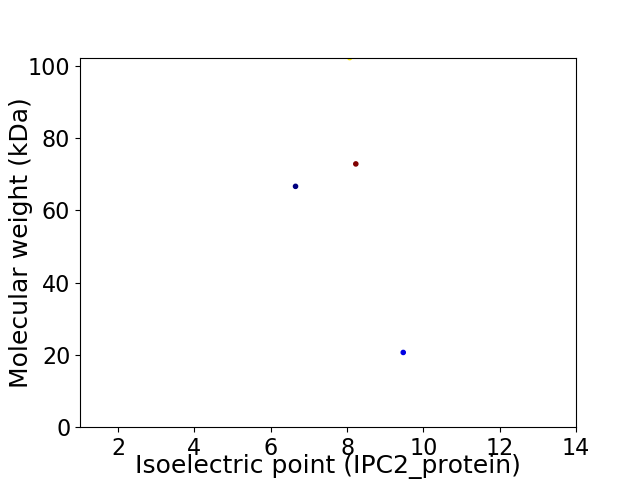
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
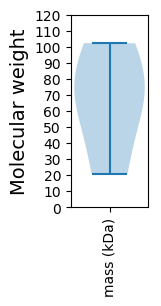
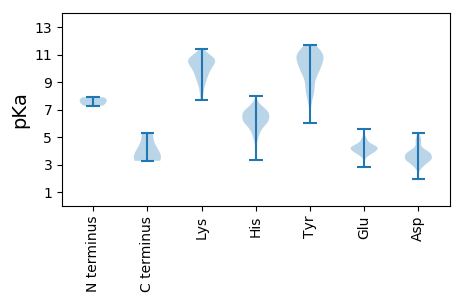
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGP6|A0A1L3KGP6_9VIRU RNA-directed RNA polymerase OS=Wuhan insect virus 21 OX=1923725 PE=4 SV=1
MM1 pKa = 7.72EE2 pKa = 5.59GPHH5 pKa = 6.29EE6 pKa = 4.46SSPRR10 pKa = 11.84HH11 pKa = 5.91CDD13 pKa = 3.39APTCEE18 pKa = 4.18QPLSEE23 pKa = 4.81GLLSPIRR30 pKa = 11.84ASVQQSYY37 pKa = 9.26GQSHH41 pKa = 6.5SDD43 pKa = 3.18YY44 pKa = 10.88SLRR47 pKa = 11.84HH48 pKa = 4.91MRR50 pKa = 11.84NHH52 pKa = 7.22GSRR55 pKa = 11.84PCQRR59 pKa = 11.84PTVRR63 pKa = 11.84FHH65 pKa = 6.36QEE67 pKa = 3.0SGRR70 pKa = 11.84RR71 pKa = 11.84PDD73 pKa = 3.41QDD75 pKa = 2.79AHH77 pKa = 7.96SPICRR82 pKa = 11.84GVPEE86 pKa = 4.78GAHH89 pKa = 6.39KK90 pKa = 10.63ADD92 pKa = 4.97SRR94 pKa = 11.84GCCHH98 pKa = 6.86GRR100 pKa = 11.84GCTGKK105 pKa = 10.58RR106 pKa = 11.84STCRR110 pKa = 11.84EE111 pKa = 3.58NHH113 pKa = 5.43VSIGHH118 pKa = 6.08HH119 pKa = 6.11PVLLHH124 pKa = 6.46HH125 pKa = 6.9PGVLDD130 pKa = 3.13MGIRR134 pKa = 11.84LLCCSTDD141 pKa = 3.44NAATATQARR150 pKa = 11.84GPPFDD155 pKa = 4.2SGCLLRR161 pKa = 11.84DD162 pKa = 3.37IRR164 pKa = 11.84DD165 pKa = 3.83DD166 pKa = 4.15GQGHH170 pKa = 6.17RR171 pKa = 11.84PHH173 pKa = 6.54RR174 pKa = 11.84TCRR177 pKa = 11.84MSTCAVLALIALTLLTISRR196 pKa = 11.84PTNCRR201 pKa = 11.84EE202 pKa = 3.51ISGATVIPGDD212 pKa = 3.43GKK214 pKa = 8.88TYY216 pKa = 10.74CSTASSGFTLRR227 pKa = 11.84TATLVSRR234 pKa = 11.84ITGKK238 pKa = 8.34HH239 pKa = 3.97TSAVLVPRR247 pKa = 11.84GLVTNDD253 pKa = 2.96KK254 pKa = 11.05SYY256 pKa = 11.25LSNDD260 pKa = 3.11RR261 pKa = 11.84FEE263 pKa = 5.13VWDD266 pKa = 3.97FDD268 pKa = 3.83KK269 pKa = 11.4QLVNTGTFISWPFEE283 pKa = 3.91HH284 pKa = 7.43PPRR287 pKa = 11.84DD288 pKa = 3.89CLTLLEE294 pKa = 4.73TNTLAHH300 pKa = 6.61SDD302 pKa = 4.03CPLSRR307 pKa = 11.84SCSNVAYY314 pKa = 10.03HH315 pKa = 6.33QLDD318 pKa = 3.82VKK320 pKa = 10.58WYY322 pKa = 8.39GKK324 pKa = 10.58EE325 pKa = 3.92SGLCTGTLLPHH336 pKa = 4.63VTVFNGTATNRR347 pKa = 11.84HH348 pKa = 5.2GTLSTDD354 pKa = 2.89WGPLYY359 pKa = 10.99SFVDD363 pKa = 3.39AGSWTDD369 pKa = 3.25VYY371 pKa = 11.62NVLPSLRR378 pKa = 11.84LLRR381 pKa = 11.84LPGRR385 pKa = 11.84DD386 pKa = 2.65IMFSYY391 pKa = 10.94DD392 pKa = 2.91ATLNLVALKK401 pKa = 10.23SDD403 pKa = 3.52PYY405 pKa = 11.13KK406 pKa = 10.58PVTVPVIIQPYY417 pKa = 7.41YY418 pKa = 11.18AEE420 pKa = 4.59GKK422 pKa = 9.45GQEE425 pKa = 3.75FCMWVNPPEE434 pKa = 4.24RR435 pKa = 11.84SRR437 pKa = 11.84ALVIYY442 pKa = 9.62DD443 pKa = 3.96GPTLTEE449 pKa = 4.27WPPFTPVLAVHH460 pKa = 7.17DD461 pKa = 5.05LDD463 pKa = 4.57TYY465 pKa = 10.94QPPICLDD472 pKa = 3.46YY473 pKa = 10.93QPRR476 pKa = 11.84PDD478 pKa = 3.61MPLNGHH484 pKa = 6.21RR485 pKa = 11.84RR486 pKa = 11.84VTMGDD491 pKa = 3.06GDD493 pKa = 3.9VGRR496 pKa = 11.84VVQDD500 pKa = 3.78TPALACAPFKK510 pKa = 10.77HH511 pKa = 6.3ISSYY515 pKa = 10.9SVPGSIWLKK524 pKa = 10.78SMLAFVIEE532 pKa = 4.52SIEE535 pKa = 4.35TVVAAAFRR543 pKa = 11.84ATITVTHH550 pKa = 6.76DD551 pKa = 4.7LIAEE555 pKa = 4.1FNDD558 pKa = 2.84RR559 pKa = 11.84FMLFEE564 pKa = 4.61ISLLAGYY571 pKa = 8.33VMWHH575 pKa = 6.04TDD577 pKa = 2.82SMIRR581 pKa = 11.84TAVVCGMYY589 pKa = 10.01TLLFGVQRR597 pKa = 11.84NPAQSPP603 pKa = 3.5
MM1 pKa = 7.72EE2 pKa = 5.59GPHH5 pKa = 6.29EE6 pKa = 4.46SSPRR10 pKa = 11.84HH11 pKa = 5.91CDD13 pKa = 3.39APTCEE18 pKa = 4.18QPLSEE23 pKa = 4.81GLLSPIRR30 pKa = 11.84ASVQQSYY37 pKa = 9.26GQSHH41 pKa = 6.5SDD43 pKa = 3.18YY44 pKa = 10.88SLRR47 pKa = 11.84HH48 pKa = 4.91MRR50 pKa = 11.84NHH52 pKa = 7.22GSRR55 pKa = 11.84PCQRR59 pKa = 11.84PTVRR63 pKa = 11.84FHH65 pKa = 6.36QEE67 pKa = 3.0SGRR70 pKa = 11.84RR71 pKa = 11.84PDD73 pKa = 3.41QDD75 pKa = 2.79AHH77 pKa = 7.96SPICRR82 pKa = 11.84GVPEE86 pKa = 4.78GAHH89 pKa = 6.39KK90 pKa = 10.63ADD92 pKa = 4.97SRR94 pKa = 11.84GCCHH98 pKa = 6.86GRR100 pKa = 11.84GCTGKK105 pKa = 10.58RR106 pKa = 11.84STCRR110 pKa = 11.84EE111 pKa = 3.58NHH113 pKa = 5.43VSIGHH118 pKa = 6.08HH119 pKa = 6.11PVLLHH124 pKa = 6.46HH125 pKa = 6.9PGVLDD130 pKa = 3.13MGIRR134 pKa = 11.84LLCCSTDD141 pKa = 3.44NAATATQARR150 pKa = 11.84GPPFDD155 pKa = 4.2SGCLLRR161 pKa = 11.84DD162 pKa = 3.37IRR164 pKa = 11.84DD165 pKa = 3.83DD166 pKa = 4.15GQGHH170 pKa = 6.17RR171 pKa = 11.84PHH173 pKa = 6.54RR174 pKa = 11.84TCRR177 pKa = 11.84MSTCAVLALIALTLLTISRR196 pKa = 11.84PTNCRR201 pKa = 11.84EE202 pKa = 3.51ISGATVIPGDD212 pKa = 3.43GKK214 pKa = 8.88TYY216 pKa = 10.74CSTASSGFTLRR227 pKa = 11.84TATLVSRR234 pKa = 11.84ITGKK238 pKa = 8.34HH239 pKa = 3.97TSAVLVPRR247 pKa = 11.84GLVTNDD253 pKa = 2.96KK254 pKa = 11.05SYY256 pKa = 11.25LSNDD260 pKa = 3.11RR261 pKa = 11.84FEE263 pKa = 5.13VWDD266 pKa = 3.97FDD268 pKa = 3.83KK269 pKa = 11.4QLVNTGTFISWPFEE283 pKa = 3.91HH284 pKa = 7.43PPRR287 pKa = 11.84DD288 pKa = 3.89CLTLLEE294 pKa = 4.73TNTLAHH300 pKa = 6.61SDD302 pKa = 4.03CPLSRR307 pKa = 11.84SCSNVAYY314 pKa = 10.03HH315 pKa = 6.33QLDD318 pKa = 3.82VKK320 pKa = 10.58WYY322 pKa = 8.39GKK324 pKa = 10.58EE325 pKa = 3.92SGLCTGTLLPHH336 pKa = 4.63VTVFNGTATNRR347 pKa = 11.84HH348 pKa = 5.2GTLSTDD354 pKa = 2.89WGPLYY359 pKa = 10.99SFVDD363 pKa = 3.39AGSWTDD369 pKa = 3.25VYY371 pKa = 11.62NVLPSLRR378 pKa = 11.84LLRR381 pKa = 11.84LPGRR385 pKa = 11.84DD386 pKa = 2.65IMFSYY391 pKa = 10.94DD392 pKa = 2.91ATLNLVALKK401 pKa = 10.23SDD403 pKa = 3.52PYY405 pKa = 11.13KK406 pKa = 10.58PVTVPVIIQPYY417 pKa = 7.41YY418 pKa = 11.18AEE420 pKa = 4.59GKK422 pKa = 9.45GQEE425 pKa = 3.75FCMWVNPPEE434 pKa = 4.24RR435 pKa = 11.84SRR437 pKa = 11.84ALVIYY442 pKa = 9.62DD443 pKa = 3.96GPTLTEE449 pKa = 4.27WPPFTPVLAVHH460 pKa = 7.17DD461 pKa = 5.05LDD463 pKa = 4.57TYY465 pKa = 10.94QPPICLDD472 pKa = 3.46YY473 pKa = 10.93QPRR476 pKa = 11.84PDD478 pKa = 3.61MPLNGHH484 pKa = 6.21RR485 pKa = 11.84RR486 pKa = 11.84VTMGDD491 pKa = 3.06GDD493 pKa = 3.9VGRR496 pKa = 11.84VVQDD500 pKa = 3.78TPALACAPFKK510 pKa = 10.77HH511 pKa = 6.3ISSYY515 pKa = 10.9SVPGSIWLKK524 pKa = 10.78SMLAFVIEE532 pKa = 4.52SIEE535 pKa = 4.35TVVAAAFRR543 pKa = 11.84ATITVTHH550 pKa = 6.76DD551 pKa = 4.7LIAEE555 pKa = 4.1FNDD558 pKa = 2.84RR559 pKa = 11.84FMLFEE564 pKa = 4.61ISLLAGYY571 pKa = 8.33VMWHH575 pKa = 6.04TDD577 pKa = 2.82SMIRR581 pKa = 11.84TAVVCGMYY589 pKa = 10.01TLLFGVQRR597 pKa = 11.84NPAQSPP603 pKa = 3.5
Molecular weight: 66.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGV1|A0A1L3KGV1_9VIRU Chropara_Vmeth domain-containing protein OS=Wuhan insect virus 21 OX=1923725 PE=4 SV=1
MM1 pKa = 7.89PRR3 pKa = 11.84RR4 pKa = 11.84QAVTSRR10 pKa = 11.84PWRR13 pKa = 11.84GLTRR17 pKa = 11.84AAPVIATRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84VSSRR31 pKa = 11.84SAKK34 pKa = 10.56DD35 pKa = 2.95FLARR39 pKa = 11.84LGQAFNRR46 pKa = 11.84VTANPIAIIAFAICAIMAVDD66 pKa = 3.69HH67 pKa = 6.98AKK69 pKa = 10.88DD70 pKa = 3.73PLSDD74 pKa = 4.11SIKK77 pKa = 10.42KK78 pKa = 9.97VADD81 pKa = 3.79ALIKK85 pKa = 10.22MPTLQFVGEE94 pKa = 4.09YY95 pKa = 9.86LKK97 pKa = 10.9EE98 pKa = 4.36HH99 pKa = 6.93IKK101 pKa = 8.68QTVGAVAMGAAVLVSARR118 pKa = 11.84PAEE121 pKa = 4.11KK122 pKa = 8.21TTYY125 pKa = 11.13LLGTTLFSYY134 pKa = 10.3IIPEE138 pKa = 3.97YY139 pKa = 10.65SIWAYY144 pKa = 10.41GCYY147 pKa = 9.72AAALTMLLQLRR158 pKa = 11.84RR159 pKa = 11.84LEE161 pKa = 4.2DD162 pKa = 3.3RR163 pKa = 11.84LLIAGACFVIYY174 pKa = 11.07VMTVKK179 pKa = 10.12DD180 pKa = 3.66TDD182 pKa = 3.08HH183 pKa = 6.64TARR186 pKa = 11.84VGG188 pKa = 3.27
MM1 pKa = 7.89PRR3 pKa = 11.84RR4 pKa = 11.84QAVTSRR10 pKa = 11.84PWRR13 pKa = 11.84GLTRR17 pKa = 11.84AAPVIATRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84VSSRR31 pKa = 11.84SAKK34 pKa = 10.56DD35 pKa = 2.95FLARR39 pKa = 11.84LGQAFNRR46 pKa = 11.84VTANPIAIIAFAICAIMAVDD66 pKa = 3.69HH67 pKa = 6.98AKK69 pKa = 10.88DD70 pKa = 3.73PLSDD74 pKa = 4.11SIKK77 pKa = 10.42KK78 pKa = 9.97VADD81 pKa = 3.79ALIKK85 pKa = 10.22MPTLQFVGEE94 pKa = 4.09YY95 pKa = 9.86LKK97 pKa = 10.9EE98 pKa = 4.36HH99 pKa = 6.93IKK101 pKa = 8.68QTVGAVAMGAAVLVSARR118 pKa = 11.84PAEE121 pKa = 4.11KK122 pKa = 8.21TTYY125 pKa = 11.13LLGTTLFSYY134 pKa = 10.3IIPEE138 pKa = 3.97YY139 pKa = 10.65SIWAYY144 pKa = 10.41GCYY147 pKa = 9.72AAALTMLLQLRR158 pKa = 11.84RR159 pKa = 11.84LEE161 pKa = 4.2DD162 pKa = 3.3RR163 pKa = 11.84LLIAGACFVIYY174 pKa = 11.07VMTVKK179 pKa = 10.12DD180 pKa = 3.66TDD182 pKa = 3.08HH183 pKa = 6.64TARR186 pKa = 11.84VGG188 pKa = 3.27
Molecular weight: 20.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2354 |
188 |
919 |
588.5 |
65.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.539 ± 0.961 | 2.421 ± 0.439 |
5.862 ± 0.403 | 4.46 ± 0.505 |
3.314 ± 0.369 | 6.415 ± 0.816 |
3.866 ± 0.587 | 5.055 ± 0.402 |
3.441 ± 0.439 | 7.986 ± 0.985 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.997 ± 0.429 | 2.974 ± 0.278 |
6.457 ± 0.394 | 3.144 ± 0.099 |
8.539 ± 0.499 | 7.264 ± 0.363 |
7.392 ± 0.347 | 6.542 ± 0.322 |
1.359 ± 0.143 | 2.974 ± 0.088 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
