
Mosquito VEM virus SDRBAJ
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|F6KIE6|F6KIE6_9VIRU Uncharacterized protein OS=Mosquito VEM virus SDRBAJ OX=1034806 PE=4 SV=1
MM1 pKa = 8.2DD2 pKa = 3.71MRR4 pKa = 11.84DD5 pKa = 3.61NPRR8 pKa = 11.84SLSMSSTDD16 pKa = 3.38GSPEE20 pKa = 3.35ISSRR24 pKa = 11.84DD25 pKa = 3.59SQIATPSLWRR35 pKa = 11.84PKK37 pKa = 10.59AEE39 pKa = 4.51PFNSWRR45 pKa = 11.84RR46 pKa = 11.84EE47 pKa = 4.07SLSPPTARR55 pKa = 11.84RR56 pKa = 11.84SPGINQDD63 pKa = 2.72WAQWDD68 pKa = 3.94EE69 pKa = 4.3DD70 pKa = 4.61FKK72 pKa = 11.45HH73 pKa = 6.19LWGTYY78 pKa = 9.8SMSGPQSAMKK88 pKa = 10.37QPIANNWMKK97 pKa = 10.87SDD99 pKa = 3.78PEE101 pKa = 4.33VIHH104 pKa = 6.48FLEE107 pKa = 4.7EE108 pKa = 3.68QWEE111 pKa = 3.97QDD113 pKa = 3.22PFEE116 pKa = 4.64VKK118 pKa = 10.36SVTWW122 pKa = 3.44
MM1 pKa = 8.2DD2 pKa = 3.71MRR4 pKa = 11.84DD5 pKa = 3.61NPRR8 pKa = 11.84SLSMSSTDD16 pKa = 3.38GSPEE20 pKa = 3.35ISSRR24 pKa = 11.84DD25 pKa = 3.59SQIATPSLWRR35 pKa = 11.84PKK37 pKa = 10.59AEE39 pKa = 4.51PFNSWRR45 pKa = 11.84RR46 pKa = 11.84EE47 pKa = 4.07SLSPPTARR55 pKa = 11.84RR56 pKa = 11.84SPGINQDD63 pKa = 2.72WAQWDD68 pKa = 3.94EE69 pKa = 4.3DD70 pKa = 4.61FKK72 pKa = 11.45HH73 pKa = 6.19LWGTYY78 pKa = 9.8SMSGPQSAMKK88 pKa = 10.37QPIANNWMKK97 pKa = 10.87SDD99 pKa = 3.78PEE101 pKa = 4.33VIHH104 pKa = 6.48FLEE107 pKa = 4.7EE108 pKa = 3.68QWEE111 pKa = 3.97QDD113 pKa = 3.22PFEE116 pKa = 4.64VKK118 pKa = 10.36SVTWW122 pKa = 3.44
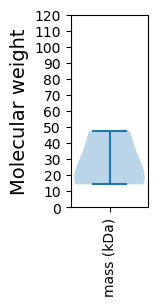
Molecular weight: 14.2 kDa
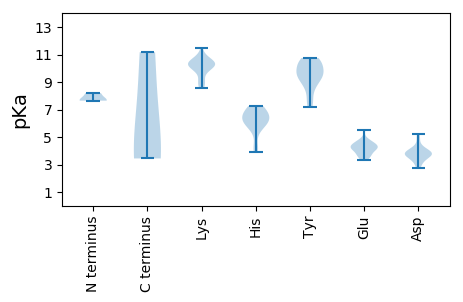
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6KIE6|F6KIE6_9VIRU Uncharacterized protein OS=Mosquito VEM virus SDRBAJ OX=1034806 PE=4 SV=1
MM1 pKa = 7.66APGKK5 pKa = 10.29KK6 pKa = 9.45SLHH9 pKa = 5.89SSEE12 pKa = 4.83SNTIRR17 pKa = 11.84EE18 pKa = 4.47HH19 pKa = 5.39YY20 pKa = 7.53QTLVVFGAGGTSLKK34 pKa = 10.8LIGEE38 pKa = 4.82IFRR41 pKa = 11.84SLYY44 pKa = 9.62SQIPIWLRR52 pKa = 11.84GIIIGKK58 pKa = 8.64YY59 pKa = 8.56PISIFLFRR67 pKa = 11.84SRR69 pKa = 11.84FRR71 pKa = 11.84HH72 pKa = 4.32NMIWEE77 pKa = 4.34GLTTGAGASGRR88 pKa = 11.84TGQRR92 pKa = 11.84LSGSRR97 pKa = 11.84EE98 pKa = 3.76GRR100 pKa = 11.84PHH102 pKa = 6.98RR103 pKa = 11.84GCGALLAHH111 pKa = 6.53GPEE114 pKa = 4.59VGGNTYY120 pKa = 9.52PHH122 pKa = 7.25LWSTHH127 pKa = 3.94TSGPRR132 pKa = 11.84ASLWANPSNKK142 pKa = 9.05INPVIHH148 pKa = 5.92YY149 pKa = 9.83VYY151 pKa = 10.24TDD153 pKa = 4.14SINDD157 pKa = 3.1NWALNLAKK165 pKa = 10.24VYY167 pKa = 9.96KK168 pKa = 10.2IPVIHH173 pKa = 7.05KK174 pKa = 8.75PRR176 pKa = 11.84SGPEE180 pKa = 3.5TTYY183 pKa = 10.04WPSGHH188 pKa = 6.52IQTMPRR194 pKa = 11.84YY195 pKa = 8.85SKK197 pKa = 9.84PIPMSINRR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84AWQAAKK213 pKa = 10.37DD214 pKa = 3.78KK215 pKa = 10.99RR216 pKa = 11.84YY217 pKa = 9.57IAYY220 pKa = 9.27GKK222 pKa = 10.26ARR224 pKa = 11.84ATVVVPGYY232 pKa = 8.88TRR234 pKa = 11.84TGGTFGRR241 pKa = 11.84FTGGPGSEE249 pKa = 3.75NKK251 pKa = 9.95FFDD254 pKa = 3.56TALGFSFDD262 pKa = 3.76STGEE266 pKa = 4.17VPATGQLNLIPQGVTEE282 pKa = 4.17STRR285 pKa = 11.84VGRR288 pKa = 11.84SCILKK293 pKa = 10.17SIQIKK298 pKa = 10.21GRR300 pKa = 11.84LRR302 pKa = 11.84FDD304 pKa = 3.67PGAAADD310 pKa = 3.98SSTIAYY316 pKa = 9.39LYY318 pKa = 10.43LVQDD322 pKa = 4.02SQCNGAAAAVTDD334 pKa = 4.11VLTSATMAGAIVNMDD349 pKa = 2.83NSQRR353 pKa = 11.84FRR355 pKa = 11.84ILKK358 pKa = 9.02KK359 pKa = 8.56WTWALNSASGVSTAYY374 pKa = 9.34NTVDD378 pKa = 3.82RR379 pKa = 11.84AIDD382 pKa = 3.78YY383 pKa = 9.48YY384 pKa = 10.47RR385 pKa = 11.84KK386 pKa = 9.79CHH388 pKa = 6.23IPLDD392 pKa = 4.27FSSTTGAIGEE402 pKa = 4.12IRR404 pKa = 11.84SNNVFLLAGSADD416 pKa = 3.75TDD418 pKa = 3.81DD419 pKa = 5.19LVAFSGVARR428 pKa = 11.84VRR430 pKa = 11.84FSDD433 pKa = 3.48KK434 pKa = 11.17
MM1 pKa = 7.66APGKK5 pKa = 10.29KK6 pKa = 9.45SLHH9 pKa = 5.89SSEE12 pKa = 4.83SNTIRR17 pKa = 11.84EE18 pKa = 4.47HH19 pKa = 5.39YY20 pKa = 7.53QTLVVFGAGGTSLKK34 pKa = 10.8LIGEE38 pKa = 4.82IFRR41 pKa = 11.84SLYY44 pKa = 9.62SQIPIWLRR52 pKa = 11.84GIIIGKK58 pKa = 8.64YY59 pKa = 8.56PISIFLFRR67 pKa = 11.84SRR69 pKa = 11.84FRR71 pKa = 11.84HH72 pKa = 4.32NMIWEE77 pKa = 4.34GLTTGAGASGRR88 pKa = 11.84TGQRR92 pKa = 11.84LSGSRR97 pKa = 11.84EE98 pKa = 3.76GRR100 pKa = 11.84PHH102 pKa = 6.98RR103 pKa = 11.84GCGALLAHH111 pKa = 6.53GPEE114 pKa = 4.59VGGNTYY120 pKa = 9.52PHH122 pKa = 7.25LWSTHH127 pKa = 3.94TSGPRR132 pKa = 11.84ASLWANPSNKK142 pKa = 9.05INPVIHH148 pKa = 5.92YY149 pKa = 9.83VYY151 pKa = 10.24TDD153 pKa = 4.14SINDD157 pKa = 3.1NWALNLAKK165 pKa = 10.24VYY167 pKa = 9.96KK168 pKa = 10.2IPVIHH173 pKa = 7.05KK174 pKa = 8.75PRR176 pKa = 11.84SGPEE180 pKa = 3.5TTYY183 pKa = 10.04WPSGHH188 pKa = 6.52IQTMPRR194 pKa = 11.84YY195 pKa = 8.85SKK197 pKa = 9.84PIPMSINRR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84AWQAAKK213 pKa = 10.37DD214 pKa = 3.78KK215 pKa = 10.99RR216 pKa = 11.84YY217 pKa = 9.57IAYY220 pKa = 9.27GKK222 pKa = 10.26ARR224 pKa = 11.84ATVVVPGYY232 pKa = 8.88TRR234 pKa = 11.84TGGTFGRR241 pKa = 11.84FTGGPGSEE249 pKa = 3.75NKK251 pKa = 9.95FFDD254 pKa = 3.56TALGFSFDD262 pKa = 3.76STGEE266 pKa = 4.17VPATGQLNLIPQGVTEE282 pKa = 4.17STRR285 pKa = 11.84VGRR288 pKa = 11.84SCILKK293 pKa = 10.17SIQIKK298 pKa = 10.21GRR300 pKa = 11.84LRR302 pKa = 11.84FDD304 pKa = 3.67PGAAADD310 pKa = 3.98SSTIAYY316 pKa = 9.39LYY318 pKa = 10.43LVQDD322 pKa = 4.02SQCNGAAAAVTDD334 pKa = 4.11VLTSATMAGAIVNMDD349 pKa = 2.83NSQRR353 pKa = 11.84FRR355 pKa = 11.84ILKK358 pKa = 9.02KK359 pKa = 8.56WTWALNSASGVSTAYY374 pKa = 9.34NTVDD378 pKa = 3.82RR379 pKa = 11.84AIDD382 pKa = 3.78YY383 pKa = 9.48YY384 pKa = 10.47RR385 pKa = 11.84KK386 pKa = 9.79CHH388 pKa = 6.23IPLDD392 pKa = 4.27FSSTTGAIGEE402 pKa = 4.12IRR404 pKa = 11.84SNNVFLLAGSADD416 pKa = 3.75TDD418 pKa = 3.81DD419 pKa = 5.19LVAFSGVARR428 pKa = 11.84VRR430 pKa = 11.84FSDD433 pKa = 3.48KK434 pKa = 11.17
Molecular weight: 47.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
725 |
122 |
434 |
241.7 |
26.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.0 ± 0.885 | 1.103 ± 0.436 |
4.414 ± 0.804 | 4.414 ± 1.585 |
3.862 ± 0.16 | 8.414 ± 1.672 |
2.621 ± 0.339 | 6.069 ± 0.915 |
5.103 ± 0.859 | 6.207 ± 0.593 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.931 ± 0.816 | 4.0 ± 0.369 |
6.069 ± 1.107 | 3.034 ± 0.739 |
6.897 ± 0.412 | 9.655 ± 1.461 |
6.483 ± 0.976 | 4.966 ± 0.678 |
2.759 ± 1.036 | 4.0 ± 0.995 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
