
Haloechinothrix alba
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Haloechinothrix
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
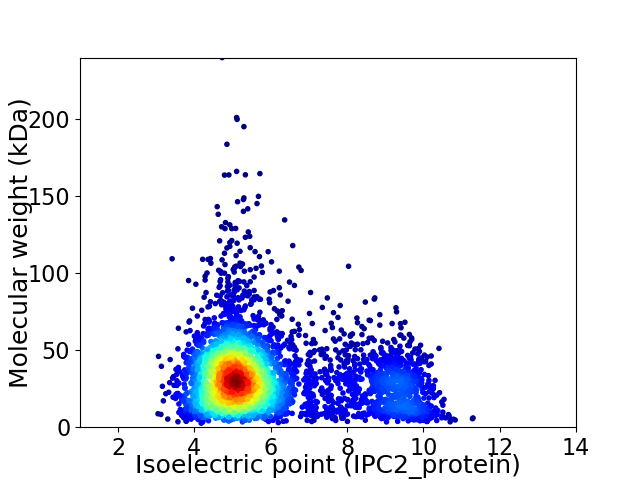
Virtual 2D-PAGE plot for 4481 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238WMW8|A0A238WMW8_9PSEU Uncharacterized protein OS=Haloechinothrix alba OX=664784 GN=SAMN06265360_1076 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84LVSRR6 pKa = 11.84RR7 pKa = 11.84VVAVLASGALATFAFAAPASASGYY31 pKa = 9.88GADD34 pKa = 4.4DD35 pKa = 4.14AGGAAINIVADD46 pKa = 4.05TVSAAQEE53 pKa = 4.27VTGSWSAEE61 pKa = 3.64RR62 pKa = 11.84MRR64 pKa = 11.84QAVPMEE70 pKa = 3.97QLVSEE75 pKa = 4.48SGDD78 pKa = 3.54VISLGEE84 pKa = 4.18VVGSLTGGDD93 pKa = 3.83PVSIPATKK101 pKa = 10.44RR102 pKa = 11.84LDD104 pKa = 3.68LGSGLGGVLGSASGEE119 pKa = 4.0PWTGGGEE126 pKa = 4.31VASTAGRR133 pKa = 11.84VFFTVDD139 pKa = 3.35GQDD142 pKa = 3.43ASCSGNAVTSEE153 pKa = 4.05NNSTVITAGHH163 pKa = 5.94CVRR166 pKa = 11.84MGGEE170 pKa = 3.71WHH172 pKa = 7.42DD173 pKa = 3.81DD174 pKa = 3.01WVFVPGYY181 pKa = 10.59DD182 pKa = 4.9NGDD185 pKa = 3.3APYY188 pKa = 10.78GEE190 pKa = 4.75WPAEE194 pKa = 3.97TTMATQEE201 pKa = 4.12WVEE204 pKa = 4.38SEE206 pKa = 4.56DD207 pKa = 3.23INYY210 pKa = 10.33DD211 pKa = 2.93VGAAVVHH218 pKa = 5.74EE219 pKa = 4.3QDD221 pKa = 3.62GQSLTEE227 pKa = 3.95VVGGQGIAFNQEE239 pKa = 3.58RR240 pKa = 11.84GQPMYY245 pKa = 11.27SFGYY249 pKa = 8.94PAASPYY255 pKa = 10.71DD256 pKa = 3.78GSEE259 pKa = 4.75LIHH262 pKa = 6.85CSGDD266 pKa = 3.53TFDD269 pKa = 6.19DD270 pKa = 4.16PLFSDD275 pKa = 5.21AMGMDD280 pKa = 4.41CDD282 pKa = 3.9MTGGSSGGPWFLDD295 pKa = 3.29FDD297 pKa = 4.27EE298 pKa = 4.79QSGTGLQSSVNSFGYY313 pKa = 10.87AFLPGTMFGPYY324 pKa = 9.89FGDD327 pKa = 3.57EE328 pKa = 3.94AQEE331 pKa = 4.22VYY333 pKa = 10.69EE334 pKa = 4.3RR335 pKa = 11.84ASTFF339 pKa = 3.1
MM1 pKa = 7.55RR2 pKa = 11.84LVSRR6 pKa = 11.84RR7 pKa = 11.84VVAVLASGALATFAFAAPASASGYY31 pKa = 9.88GADD34 pKa = 4.4DD35 pKa = 4.14AGGAAINIVADD46 pKa = 4.05TVSAAQEE53 pKa = 4.27VTGSWSAEE61 pKa = 3.64RR62 pKa = 11.84MRR64 pKa = 11.84QAVPMEE70 pKa = 3.97QLVSEE75 pKa = 4.48SGDD78 pKa = 3.54VISLGEE84 pKa = 4.18VVGSLTGGDD93 pKa = 3.83PVSIPATKK101 pKa = 10.44RR102 pKa = 11.84LDD104 pKa = 3.68LGSGLGGVLGSASGEE119 pKa = 4.0PWTGGGEE126 pKa = 4.31VASTAGRR133 pKa = 11.84VFFTVDD139 pKa = 3.35GQDD142 pKa = 3.43ASCSGNAVTSEE153 pKa = 4.05NNSTVITAGHH163 pKa = 5.94CVRR166 pKa = 11.84MGGEE170 pKa = 3.71WHH172 pKa = 7.42DD173 pKa = 3.81DD174 pKa = 3.01WVFVPGYY181 pKa = 10.59DD182 pKa = 4.9NGDD185 pKa = 3.3APYY188 pKa = 10.78GEE190 pKa = 4.75WPAEE194 pKa = 3.97TTMATQEE201 pKa = 4.12WVEE204 pKa = 4.38SEE206 pKa = 4.56DD207 pKa = 3.23INYY210 pKa = 10.33DD211 pKa = 2.93VGAAVVHH218 pKa = 5.74EE219 pKa = 4.3QDD221 pKa = 3.62GQSLTEE227 pKa = 3.95VVGGQGIAFNQEE239 pKa = 3.58RR240 pKa = 11.84GQPMYY245 pKa = 11.27SFGYY249 pKa = 8.94PAASPYY255 pKa = 10.71DD256 pKa = 3.78GSEE259 pKa = 4.75LIHH262 pKa = 6.85CSGDD266 pKa = 3.53TFDD269 pKa = 6.19DD270 pKa = 4.16PLFSDD275 pKa = 5.21AMGMDD280 pKa = 4.41CDD282 pKa = 3.9MTGGSSGGPWFLDD295 pKa = 3.29FDD297 pKa = 4.27EE298 pKa = 4.79QSGTGLQSSVNSFGYY313 pKa = 10.87AFLPGTMFGPYY324 pKa = 9.89FGDD327 pKa = 3.57EE328 pKa = 3.94AQEE331 pKa = 4.22VYY333 pKa = 10.69EE334 pKa = 4.3RR335 pKa = 11.84ASTFF339 pKa = 3.1
Molecular weight: 35.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239AWC3|A0A239AWC3_9PSEU Integrase core domain-containing protein (Fragment) OS=Haloechinothrix alba OX=664784 GN=SAMN06265360_1614 PE=4 SV=1
MM1 pKa = 7.85RR2 pKa = 11.84FARR5 pKa = 11.84PKK7 pKa = 10.63AGVALGRR14 pKa = 11.84GLGALGRR21 pKa = 11.84WVRR24 pKa = 11.84RR25 pKa = 11.84VRR27 pKa = 11.84LPRR30 pKa = 11.84VRR32 pKa = 11.84VGVLWGWLRR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84VSGRR48 pKa = 11.84GLAAVAA54 pKa = 4.27
MM1 pKa = 7.85RR2 pKa = 11.84FARR5 pKa = 11.84PKK7 pKa = 10.63AGVALGRR14 pKa = 11.84GLGALGRR21 pKa = 11.84WVRR24 pKa = 11.84RR25 pKa = 11.84VRR27 pKa = 11.84LPRR30 pKa = 11.84VRR32 pKa = 11.84VGVLWGWLRR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84VSGRR48 pKa = 11.84GLAAVAA54 pKa = 4.27
Molecular weight: 5.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1395595 |
25 |
2307 |
311.4 |
33.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.665 ± 0.052 | 0.847 ± 0.01 |
6.395 ± 0.029 | 6.569 ± 0.035 |
2.697 ± 0.02 | 9.335 ± 0.033 |
2.407 ± 0.017 | 3.519 ± 0.022 |
1.784 ± 0.024 | 9.821 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.888 ± 0.016 | 1.867 ± 0.016 |
5.544 ± 0.031 | 2.873 ± 0.021 |
8.064 ± 0.033 | 5.545 ± 0.024 |
5.972 ± 0.023 | 8.743 ± 0.034 |
1.407 ± 0.016 | 2.058 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
