
Hubei mosquito virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
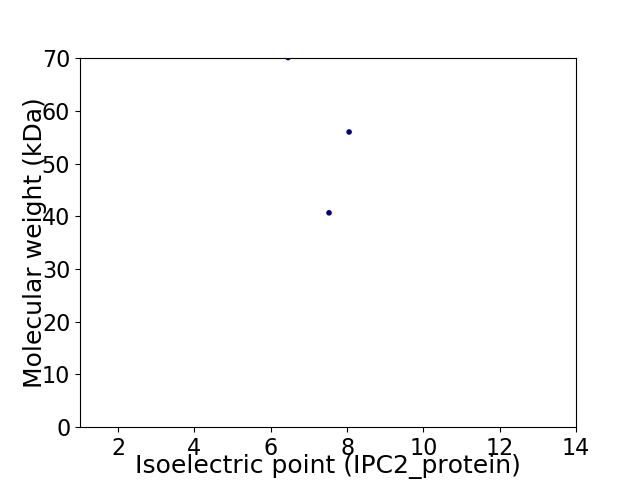
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
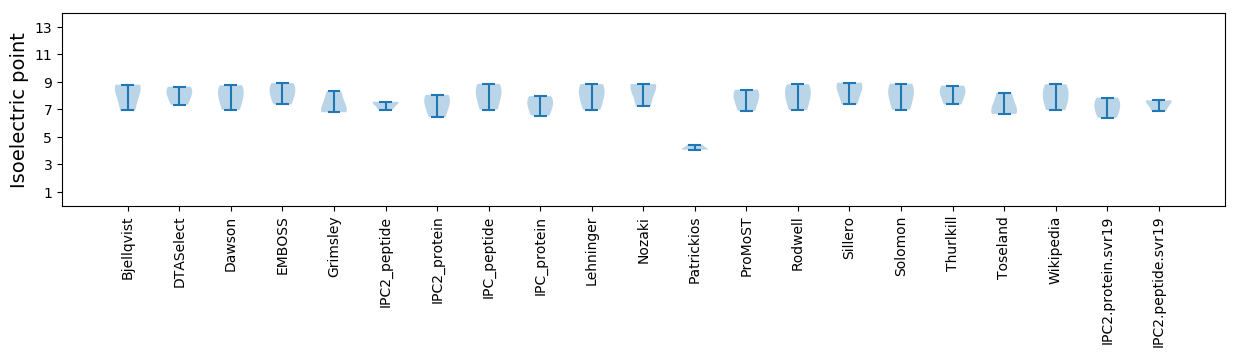
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEJ4|A0A1L3KEJ4_9VIRU RNA-directed RNA polymerase OS=Hubei mosquito virus 2 OX=1922926 PE=4 SV=1
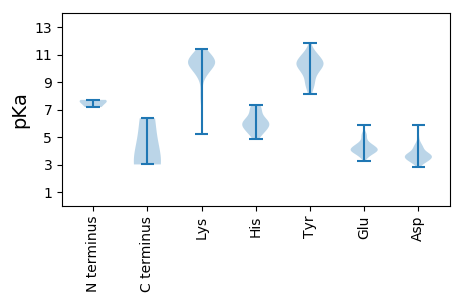
MM1 pKa = 7.18YY2 pKa = 10.23KK3 pKa = 9.94PYY5 pKa = 10.7TMIMEE10 pKa = 4.93DD11 pKa = 4.34LFISHH16 pKa = 6.61YY17 pKa = 11.59AMLVYY22 pKa = 10.33VALLVFLALTIVNVINPCTKK42 pKa = 10.32AVCTCSFAAVFVMFCRR58 pKa = 11.84ALALVTRR65 pKa = 11.84QFLFQGIHH73 pKa = 5.22GVVVVLTSKK82 pKa = 10.81KK83 pKa = 8.33FWQSFYY89 pKa = 10.34VTTPEE94 pKa = 4.15IFYY97 pKa = 10.76TMIAFYY103 pKa = 9.39LTLALVIAFNVWYY116 pKa = 9.98KK117 pKa = 10.36RR118 pKa = 11.84RR119 pKa = 11.84EE120 pKa = 4.16AMRR123 pKa = 11.84HH124 pKa = 4.83KK125 pKa = 10.99YY126 pKa = 8.92MVLNEE131 pKa = 4.16SGTYY135 pKa = 8.66MEE137 pKa = 5.18EE138 pKa = 3.97KK139 pKa = 10.73AMPNSHH145 pKa = 6.05YY146 pKa = 10.41EE147 pKa = 3.95IVPVLPGFQAAVLVSLDD164 pKa = 3.64GITYY168 pKa = 10.16RR169 pKa = 11.84EE170 pKa = 3.94NGQCFWVDD178 pKa = 3.29EE179 pKa = 4.46GLVTAAHH186 pKa = 5.37VVEE189 pKa = 5.87GYY191 pKa = 10.28DD192 pKa = 3.22YY193 pKa = 11.03CCIYY197 pKa = 10.52RR198 pKa = 11.84DD199 pKa = 3.81EE200 pKa = 4.17KK201 pKa = 11.24HH202 pKa = 6.54KK203 pKa = 10.97IEE205 pKa = 4.33VPSSVFEE212 pKa = 4.2TGQGDD217 pKa = 4.16YY218 pKa = 10.9ACCRR222 pKa = 11.84DD223 pKa = 4.43PIKK226 pKa = 10.1ITQKK230 pKa = 10.63VGLSKK235 pKa = 11.09AKK237 pKa = 10.18LAGVGLEE244 pKa = 4.05KK245 pKa = 10.94GSGISANIVARR256 pKa = 11.84GKK258 pKa = 10.58RR259 pKa = 11.84SIGFLDD265 pKa = 3.15QHH267 pKa = 5.93PQFGFVTYY275 pKa = 10.44SGSTTSGFSGAPYY288 pKa = 10.21HH289 pKa = 7.03LGRR292 pKa = 11.84TILGMHH298 pKa = 7.14LGADD302 pKa = 4.17SVNMGYY308 pKa = 9.9EE309 pKa = 3.71AAFLKK314 pKa = 10.33TEE316 pKa = 4.24LRR318 pKa = 11.84PSRR321 pKa = 11.84VIKK324 pKa = 10.68KK325 pKa = 10.32LVDD328 pKa = 3.84LKK330 pKa = 11.32KK331 pKa = 10.43EE332 pKa = 4.4DD333 pKa = 3.59SASWLVEE340 pKa = 3.53HH341 pKa = 7.33LGRR344 pKa = 11.84EE345 pKa = 4.18DD346 pKa = 4.85EE347 pKa = 4.17IQYY350 pKa = 8.13WQSPFDD356 pKa = 3.79PSEE359 pKa = 4.06YY360 pKa = 10.1KK361 pKa = 10.8LKK363 pKa = 10.35IGNMYY368 pKa = 10.08HH369 pKa = 7.09IVDD372 pKa = 3.8NEE374 pKa = 4.0VLKK377 pKa = 11.05EE378 pKa = 3.48ILTRR382 pKa = 11.84KK383 pKa = 9.62KK384 pKa = 10.47GRR386 pKa = 11.84QAVDD390 pKa = 3.19KK391 pKa = 10.48IDD393 pKa = 3.58YY394 pKa = 10.47LEE396 pKa = 4.83EE397 pKa = 5.31SDD399 pKa = 5.4APRR402 pKa = 11.84KK403 pKa = 5.21TTKK406 pKa = 9.52STEE409 pKa = 4.04TMVWRR414 pKa = 11.84EE415 pKa = 3.85DD416 pKa = 3.37VEE418 pKa = 4.37EE419 pKa = 4.11SVRR422 pKa = 11.84VYY424 pKa = 10.84GGEE427 pKa = 3.64VDD429 pKa = 5.62LFDD432 pKa = 4.49EE433 pKa = 4.63PVIVPVPVEE442 pKa = 4.43DD443 pKa = 3.39RR444 pKa = 11.84HH445 pKa = 6.11EE446 pKa = 4.72DD447 pKa = 3.58PLIATIRR454 pKa = 11.84GLVQEE459 pKa = 5.02FGRR462 pKa = 11.84TSVYY466 pKa = 10.44TSAQLPAVKK475 pKa = 9.69EE476 pKa = 4.05AAQVPPVKK484 pKa = 9.98EE485 pKa = 4.0AEE487 pKa = 4.33AVVPAAPRR495 pKa = 11.84VDD497 pKa = 4.3EE498 pKa = 5.32LPLAPRR504 pKa = 11.84DD505 pKa = 3.39AMTFNDD511 pKa = 3.58SGNLFRR517 pKa = 11.84APAVPAGAPGMVSVPAPVQNQGQPIYY543 pKa = 11.11GPMAFPYY550 pKa = 9.79PPPLVNYY557 pKa = 9.67HH558 pKa = 5.43MEE560 pKa = 4.27SRR562 pKa = 11.84SSMLAPQNVAFTRR575 pKa = 11.84TQRR578 pKa = 11.84NRR580 pKa = 11.84NRR582 pKa = 11.84RR583 pKa = 11.84AARR586 pKa = 11.84QQKK589 pKa = 9.87RR590 pKa = 11.84SEE592 pKa = 4.07LEE594 pKa = 3.53QYY596 pKa = 9.67RR597 pKa = 11.84QRR599 pKa = 11.84FGPLQNGGATSQQPPTPTHH618 pKa = 6.26GSIEE622 pKa = 4.42SSTGQQ627 pKa = 3.09
MM1 pKa = 7.18YY2 pKa = 10.23KK3 pKa = 9.94PYY5 pKa = 10.7TMIMEE10 pKa = 4.93DD11 pKa = 4.34LFISHH16 pKa = 6.61YY17 pKa = 11.59AMLVYY22 pKa = 10.33VALLVFLALTIVNVINPCTKK42 pKa = 10.32AVCTCSFAAVFVMFCRR58 pKa = 11.84ALALVTRR65 pKa = 11.84QFLFQGIHH73 pKa = 5.22GVVVVLTSKK82 pKa = 10.81KK83 pKa = 8.33FWQSFYY89 pKa = 10.34VTTPEE94 pKa = 4.15IFYY97 pKa = 10.76TMIAFYY103 pKa = 9.39LTLALVIAFNVWYY116 pKa = 9.98KK117 pKa = 10.36RR118 pKa = 11.84RR119 pKa = 11.84EE120 pKa = 4.16AMRR123 pKa = 11.84HH124 pKa = 4.83KK125 pKa = 10.99YY126 pKa = 8.92MVLNEE131 pKa = 4.16SGTYY135 pKa = 8.66MEE137 pKa = 5.18EE138 pKa = 3.97KK139 pKa = 10.73AMPNSHH145 pKa = 6.05YY146 pKa = 10.41EE147 pKa = 3.95IVPVLPGFQAAVLVSLDD164 pKa = 3.64GITYY168 pKa = 10.16RR169 pKa = 11.84EE170 pKa = 3.94NGQCFWVDD178 pKa = 3.29EE179 pKa = 4.46GLVTAAHH186 pKa = 5.37VVEE189 pKa = 5.87GYY191 pKa = 10.28DD192 pKa = 3.22YY193 pKa = 11.03CCIYY197 pKa = 10.52RR198 pKa = 11.84DD199 pKa = 3.81EE200 pKa = 4.17KK201 pKa = 11.24HH202 pKa = 6.54KK203 pKa = 10.97IEE205 pKa = 4.33VPSSVFEE212 pKa = 4.2TGQGDD217 pKa = 4.16YY218 pKa = 10.9ACCRR222 pKa = 11.84DD223 pKa = 4.43PIKK226 pKa = 10.1ITQKK230 pKa = 10.63VGLSKK235 pKa = 11.09AKK237 pKa = 10.18LAGVGLEE244 pKa = 4.05KK245 pKa = 10.94GSGISANIVARR256 pKa = 11.84GKK258 pKa = 10.58RR259 pKa = 11.84SIGFLDD265 pKa = 3.15QHH267 pKa = 5.93PQFGFVTYY275 pKa = 10.44SGSTTSGFSGAPYY288 pKa = 10.21HH289 pKa = 7.03LGRR292 pKa = 11.84TILGMHH298 pKa = 7.14LGADD302 pKa = 4.17SVNMGYY308 pKa = 9.9EE309 pKa = 3.71AAFLKK314 pKa = 10.33TEE316 pKa = 4.24LRR318 pKa = 11.84PSRR321 pKa = 11.84VIKK324 pKa = 10.68KK325 pKa = 10.32LVDD328 pKa = 3.84LKK330 pKa = 11.32KK331 pKa = 10.43EE332 pKa = 4.4DD333 pKa = 3.59SASWLVEE340 pKa = 3.53HH341 pKa = 7.33LGRR344 pKa = 11.84EE345 pKa = 4.18DD346 pKa = 4.85EE347 pKa = 4.17IQYY350 pKa = 8.13WQSPFDD356 pKa = 3.79PSEE359 pKa = 4.06YY360 pKa = 10.1KK361 pKa = 10.8LKK363 pKa = 10.35IGNMYY368 pKa = 10.08HH369 pKa = 7.09IVDD372 pKa = 3.8NEE374 pKa = 4.0VLKK377 pKa = 11.05EE378 pKa = 3.48ILTRR382 pKa = 11.84KK383 pKa = 9.62KK384 pKa = 10.47GRR386 pKa = 11.84QAVDD390 pKa = 3.19KK391 pKa = 10.48IDD393 pKa = 3.58YY394 pKa = 10.47LEE396 pKa = 4.83EE397 pKa = 5.31SDD399 pKa = 5.4APRR402 pKa = 11.84KK403 pKa = 5.21TTKK406 pKa = 9.52STEE409 pKa = 4.04TMVWRR414 pKa = 11.84EE415 pKa = 3.85DD416 pKa = 3.37VEE418 pKa = 4.37EE419 pKa = 4.11SVRR422 pKa = 11.84VYY424 pKa = 10.84GGEE427 pKa = 3.64VDD429 pKa = 5.62LFDD432 pKa = 4.49EE433 pKa = 4.63PVIVPVPVEE442 pKa = 4.43DD443 pKa = 3.39RR444 pKa = 11.84HH445 pKa = 6.11EE446 pKa = 4.72DD447 pKa = 3.58PLIATIRR454 pKa = 11.84GLVQEE459 pKa = 5.02FGRR462 pKa = 11.84TSVYY466 pKa = 10.44TSAQLPAVKK475 pKa = 9.69EE476 pKa = 4.05AAQVPPVKK484 pKa = 9.98EE485 pKa = 4.0AEE487 pKa = 4.33AVVPAAPRR495 pKa = 11.84VDD497 pKa = 4.3EE498 pKa = 5.32LPLAPRR504 pKa = 11.84DD505 pKa = 3.39AMTFNDD511 pKa = 3.58SGNLFRR517 pKa = 11.84APAVPAGAPGMVSVPAPVQNQGQPIYY543 pKa = 11.11GPMAFPYY550 pKa = 9.79PPPLVNYY557 pKa = 9.67HH558 pKa = 5.43MEE560 pKa = 4.27SRR562 pKa = 11.84SSMLAPQNVAFTRR575 pKa = 11.84TQRR578 pKa = 11.84NRR580 pKa = 11.84NRR582 pKa = 11.84RR583 pKa = 11.84AARR586 pKa = 11.84QQKK589 pKa = 9.87RR590 pKa = 11.84SEE592 pKa = 4.07LEE594 pKa = 3.53QYY596 pKa = 9.67RR597 pKa = 11.84QRR599 pKa = 11.84FGPLQNGGATSQQPPTPTHH618 pKa = 6.26GSIEE622 pKa = 4.42SSTGQQ627 pKa = 3.09
Molecular weight: 70.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEG1|A0A1L3KEG1_9VIRU Uncharacterized protein OS=Hubei mosquito virus 2 OX=1922926 PE=4 SV=1
MM1 pKa = 7.57SKK3 pKa = 9.99QLKK6 pKa = 9.15KK7 pKa = 10.56QLNALTAQVAALKK20 pKa = 10.37VGKK23 pKa = 8.41QQQPAPAKK31 pKa = 9.68RR32 pKa = 11.84KK33 pKa = 9.38RR34 pKa = 11.84NRR36 pKa = 11.84KK37 pKa = 9.06RR38 pKa = 11.84KK39 pKa = 9.65GPSPLSNTGISSDD52 pKa = 3.0GRR54 pKa = 11.84IVVSRR59 pKa = 11.84CEE61 pKa = 3.82LLFEE65 pKa = 4.42VTVDD69 pKa = 3.43AAKK72 pKa = 10.75DD73 pKa = 3.42SAAFYY78 pKa = 10.96YY79 pKa = 10.35VLQASATRR87 pKa = 11.84MPWLSKK93 pKa = 9.63LAKK96 pKa = 10.07NFSQIVWRR104 pKa = 11.84SARR107 pKa = 11.84LEE109 pKa = 3.8YY110 pKa = 10.48RR111 pKa = 11.84PAVGTMKK118 pKa = 10.68DD119 pKa = 3.22GSLVIGIDD127 pKa = 3.53WNPVASAPDD136 pKa = 3.52KK137 pKa = 11.42AKK139 pKa = 10.41VQSMTPNFQVPVWQKK154 pKa = 11.19KK155 pKa = 8.07EE156 pKa = 3.8LTIPSQRR163 pKa = 11.84LMSRR167 pKa = 11.84KK168 pKa = 9.7YY169 pKa = 10.26YY170 pKa = 9.94STDD173 pKa = 2.78SSSNVQLVDD182 pKa = 3.66KK183 pKa = 10.03VPCEE187 pKa = 3.75VLGYY191 pKa = 9.37LSCTADD197 pKa = 4.34DD198 pKa = 4.16KK199 pKa = 11.37NKK201 pKa = 10.52QYY203 pKa = 10.97FGDD206 pKa = 3.29IWIHH210 pKa = 5.69YY211 pKa = 8.91NVILQGPQXGSEE223 pKa = 4.46EE224 pKa = 4.56PPPPPPPPPPPPAPTVFNYY243 pKa = 10.57NVVRR247 pKa = 11.84DD248 pKa = 3.81VTQYY252 pKa = 10.56EE253 pKa = 3.91IRR255 pKa = 11.84GSVRR259 pKa = 11.84TIFKK263 pKa = 10.84DD264 pKa = 3.5RR265 pKa = 11.84MAPYY269 pKa = 9.84PYY271 pKa = 8.8DD272 pKa = 3.64TNVFWPEE279 pKa = 3.29EE280 pKa = 4.03TVILNRR286 pKa = 11.84DD287 pKa = 3.65LIEE290 pKa = 4.08YY291 pKa = 10.12SGSDD295 pKa = 3.15NVAVNGKK302 pKa = 9.6DD303 pKa = 3.45FNYY306 pKa = 9.61VDD308 pKa = 4.06QYY310 pKa = 11.84AVSADD315 pKa = 3.84RR316 pKa = 11.84QEE318 pKa = 5.2DD319 pKa = 3.69GHH321 pKa = 6.03EE322 pKa = 4.26TTLEE326 pKa = 4.59LISRR330 pKa = 11.84PPYY333 pKa = 9.64SDD335 pKa = 2.92KK336 pKa = 11.06VPYY339 pKa = 10.07GVLDD343 pKa = 3.2ITISIRR349 pKa = 11.84SPHH352 pKa = 5.94FPRR355 pKa = 11.84LAMYY359 pKa = 8.86EE360 pKa = 4.09CAIQFVADD368 pKa = 4.21RR369 pKa = 11.84NIFDD373 pKa = 3.74SQQIRR378 pKa = 11.84QIDD381 pKa = 3.64VAFDD385 pKa = 3.19KK386 pKa = 10.79TKK388 pKa = 10.44EE389 pKa = 3.7IRR391 pKa = 11.84LVGDD395 pKa = 3.08VLNVYY400 pKa = 8.11RR401 pKa = 11.84TTNRR405 pKa = 11.84RR406 pKa = 11.84LDD408 pKa = 3.31NWYY411 pKa = 9.06MIKK414 pKa = 10.22FCQVLEE420 pKa = 4.09ILDD423 pKa = 3.54PRR425 pKa = 11.84VAVRR429 pKa = 11.84VRR431 pKa = 11.84VRR433 pKa = 11.84INRR436 pKa = 11.84LGKK439 pKa = 10.12HH440 pKa = 5.92NDD442 pKa = 3.52QIDD445 pKa = 3.55MGTKK449 pKa = 10.18LDD451 pKa = 3.76ITPVLSWYY459 pKa = 9.68TSVTKK464 pKa = 10.24FLGVGEE470 pKa = 4.55GSDD473 pKa = 3.59DD474 pKa = 3.7GSRR477 pKa = 11.84SPEE480 pKa = 3.84LEE482 pKa = 3.61GSYY485 pKa = 10.43EE486 pKa = 4.08ILDD489 pKa = 4.49DD490 pKa = 4.58PPPSGSSS497 pKa = 3.01
MM1 pKa = 7.57SKK3 pKa = 9.99QLKK6 pKa = 9.15KK7 pKa = 10.56QLNALTAQVAALKK20 pKa = 10.37VGKK23 pKa = 8.41QQQPAPAKK31 pKa = 9.68RR32 pKa = 11.84KK33 pKa = 9.38RR34 pKa = 11.84NRR36 pKa = 11.84KK37 pKa = 9.06RR38 pKa = 11.84KK39 pKa = 9.65GPSPLSNTGISSDD52 pKa = 3.0GRR54 pKa = 11.84IVVSRR59 pKa = 11.84CEE61 pKa = 3.82LLFEE65 pKa = 4.42VTVDD69 pKa = 3.43AAKK72 pKa = 10.75DD73 pKa = 3.42SAAFYY78 pKa = 10.96YY79 pKa = 10.35VLQASATRR87 pKa = 11.84MPWLSKK93 pKa = 9.63LAKK96 pKa = 10.07NFSQIVWRR104 pKa = 11.84SARR107 pKa = 11.84LEE109 pKa = 3.8YY110 pKa = 10.48RR111 pKa = 11.84PAVGTMKK118 pKa = 10.68DD119 pKa = 3.22GSLVIGIDD127 pKa = 3.53WNPVASAPDD136 pKa = 3.52KK137 pKa = 11.42AKK139 pKa = 10.41VQSMTPNFQVPVWQKK154 pKa = 11.19KK155 pKa = 8.07EE156 pKa = 3.8LTIPSQRR163 pKa = 11.84LMSRR167 pKa = 11.84KK168 pKa = 9.7YY169 pKa = 10.26YY170 pKa = 9.94STDD173 pKa = 2.78SSSNVQLVDD182 pKa = 3.66KK183 pKa = 10.03VPCEE187 pKa = 3.75VLGYY191 pKa = 9.37LSCTADD197 pKa = 4.34DD198 pKa = 4.16KK199 pKa = 11.37NKK201 pKa = 10.52QYY203 pKa = 10.97FGDD206 pKa = 3.29IWIHH210 pKa = 5.69YY211 pKa = 8.91NVILQGPQXGSEE223 pKa = 4.46EE224 pKa = 4.56PPPPPPPPPPPPAPTVFNYY243 pKa = 10.57NVVRR247 pKa = 11.84DD248 pKa = 3.81VTQYY252 pKa = 10.56EE253 pKa = 3.91IRR255 pKa = 11.84GSVRR259 pKa = 11.84TIFKK263 pKa = 10.84DD264 pKa = 3.5RR265 pKa = 11.84MAPYY269 pKa = 9.84PYY271 pKa = 8.8DD272 pKa = 3.64TNVFWPEE279 pKa = 3.29EE280 pKa = 4.03TVILNRR286 pKa = 11.84DD287 pKa = 3.65LIEE290 pKa = 4.08YY291 pKa = 10.12SGSDD295 pKa = 3.15NVAVNGKK302 pKa = 9.6DD303 pKa = 3.45FNYY306 pKa = 9.61VDD308 pKa = 4.06QYY310 pKa = 11.84AVSADD315 pKa = 3.84RR316 pKa = 11.84QEE318 pKa = 5.2DD319 pKa = 3.69GHH321 pKa = 6.03EE322 pKa = 4.26TTLEE326 pKa = 4.59LISRR330 pKa = 11.84PPYY333 pKa = 9.64SDD335 pKa = 2.92KK336 pKa = 11.06VPYY339 pKa = 10.07GVLDD343 pKa = 3.2ITISIRR349 pKa = 11.84SPHH352 pKa = 5.94FPRR355 pKa = 11.84LAMYY359 pKa = 8.86EE360 pKa = 4.09CAIQFVADD368 pKa = 4.21RR369 pKa = 11.84NIFDD373 pKa = 3.74SQQIRR378 pKa = 11.84QIDD381 pKa = 3.64VAFDD385 pKa = 3.19KK386 pKa = 10.79TKK388 pKa = 10.44EE389 pKa = 3.7IRR391 pKa = 11.84LVGDD395 pKa = 3.08VLNVYY400 pKa = 8.11RR401 pKa = 11.84TTNRR405 pKa = 11.84RR406 pKa = 11.84LDD408 pKa = 3.31NWYY411 pKa = 9.06MIKK414 pKa = 10.22FCQVLEE420 pKa = 4.09ILDD423 pKa = 3.54PRR425 pKa = 11.84VAVRR429 pKa = 11.84VRR431 pKa = 11.84VRR433 pKa = 11.84INRR436 pKa = 11.84LGKK439 pKa = 10.12HH440 pKa = 5.92NDD442 pKa = 3.52QIDD445 pKa = 3.55MGTKK449 pKa = 10.18LDD451 pKa = 3.76ITPVLSWYY459 pKa = 9.68TSVTKK464 pKa = 10.24FLGVGEE470 pKa = 4.55GSDD473 pKa = 3.59DD474 pKa = 3.7GSRR477 pKa = 11.84SPEE480 pKa = 3.84LEE482 pKa = 3.61GSYY485 pKa = 10.43EE486 pKa = 4.08ILDD489 pKa = 4.49DD490 pKa = 4.58PPPSGSSS497 pKa = 3.01
Molecular weight: 55.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1475 |
351 |
627 |
491.7 |
55.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.644 ± 0.753 | 1.559 ± 0.32 |
5.492 ± 0.824 | 5.288 ± 0.705 |
4.203 ± 0.507 | 6.034 ± 0.423 |
1.559 ± 0.363 | 4.949 ± 0.276 |
5.356 ± 0.281 | 8.0 ± 0.772 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.508 ± 0.306 | 3.593 ± 0.394 |
6.712 ± 0.601 | 4.881 ± 0.219 |
6.034 ± 0.266 | 6.508 ± 0.64 |
5.017 ± 0.3 | 9.288 ± 0.223 |
1.831 ± 0.598 | 4.475 ± 0.08 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
