
Stigmatella aurantiaca (strain DW4/3-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Archangiaceae; Stigmatella; Stigmatella aurantiaca
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
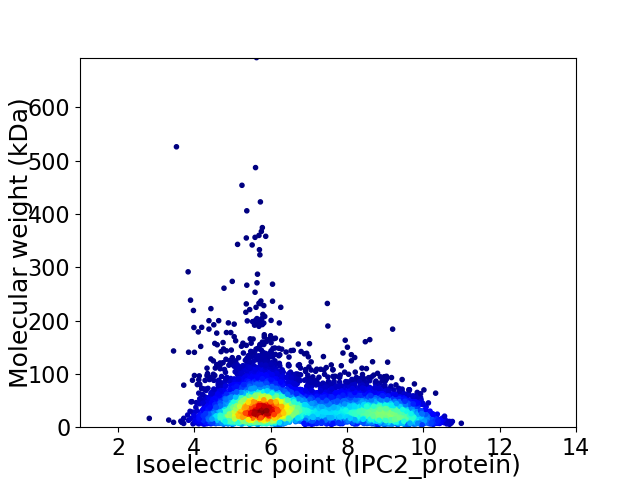
Virtual 2D-PAGE plot for 8307 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q098Q7|Q098Q7_STIAD Alpha-D-phosphohexomutase OS=Stigmatella aurantiaca (strain DW4/3-1) OX=378806 GN=STAUR_6378 PE=3 SV=1
MM1 pKa = 7.89RR2 pKa = 11.84FLRR5 pKa = 11.84SFWVLAALALTACSLLVDD23 pKa = 5.25FDD25 pKa = 6.11AEE27 pKa = 4.27GQPCDD32 pKa = 4.49SQNQCLAGYY41 pKa = 8.81VCEE44 pKa = 4.7NGSCVSGEE52 pKa = 4.15APPGGDD58 pKa = 3.52GGLGGPDD65 pKa = 3.02GGAGADD71 pKa = 3.61AGPGAGRR78 pKa = 11.84EE79 pKa = 4.16DD80 pKa = 3.92AGPDD84 pKa = 3.15AGADD88 pKa = 3.53GGDD91 pKa = 3.6GGRR94 pKa = 4.12
MM1 pKa = 7.89RR2 pKa = 11.84FLRR5 pKa = 11.84SFWVLAALALTACSLLVDD23 pKa = 5.25FDD25 pKa = 6.11AEE27 pKa = 4.27GQPCDD32 pKa = 4.49SQNQCLAGYY41 pKa = 8.81VCEE44 pKa = 4.7NGSCVSGEE52 pKa = 4.15APPGGDD58 pKa = 3.52GGLGGPDD65 pKa = 3.02GGAGADD71 pKa = 3.61AGPGAGRR78 pKa = 11.84EE79 pKa = 4.16DD80 pKa = 3.92AGPDD84 pKa = 3.15AGADD88 pKa = 3.53GGDD91 pKa = 3.6GGRR94 pKa = 4.12
Molecular weight: 9.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3FWM3|E3FWM3_STIAD Uncharacterized protein OS=Stigmatella aurantiaca (strain DW4/3-1) OX=378806 GN=STAUR_0732 PE=4 SV=1
MM1 pKa = 7.64PWHH4 pKa = 6.72EE5 pKa = 4.57PVPHH9 pKa = 7.3RR10 pKa = 11.84PRR12 pKa = 11.84QRR14 pKa = 11.84AAPAFSLPGKK24 pKa = 9.16RR25 pKa = 11.84ACGTLRR31 pKa = 11.84PTIRR35 pKa = 11.84LLFSILFWSFFALSSLVFYY54 pKa = 11.03LGALVLWALTLPFDD68 pKa = 4.05RR69 pKa = 11.84NGRR72 pKa = 11.84ILHH75 pKa = 6.84LYY77 pKa = 8.69SCFWAQLYY85 pKa = 10.31IYY87 pKa = 9.82VNPLWSSRR95 pKa = 11.84VEE97 pKa = 4.07GRR99 pKa = 11.84EE100 pKa = 3.69NLPWRR105 pKa = 11.84GAAVLVANHH114 pKa = 6.58EE115 pKa = 4.46SLGDD119 pKa = 3.57ILVLFGLYY127 pKa = 10.06RR128 pKa = 11.84PFKK131 pKa = 9.35WVSKK135 pKa = 9.85AANFRR140 pKa = 11.84LPFIGWNMTLNRR152 pKa = 11.84YY153 pKa = 8.95VPLVRR158 pKa = 11.84GDD160 pKa = 3.64RR161 pKa = 11.84EE162 pKa = 4.35SVGRR166 pKa = 11.84MMVEE170 pKa = 3.85CEE172 pKa = 3.36KK173 pKa = 10.43WLLRR177 pKa = 11.84GVPILMFPEE186 pKa = 5.23GTRR189 pKa = 11.84SPDD192 pKa = 3.21GNIKK196 pKa = 10.35AFKK199 pKa = 10.66DD200 pKa = 3.68GAFQLAVKK208 pKa = 8.0LQCPVIPVVLTGTARR223 pKa = 11.84TLPKK227 pKa = 10.29HH228 pKa = 5.06GLKK231 pKa = 10.56LQTTARR237 pKa = 11.84CRR239 pKa = 11.84VRR241 pKa = 11.84VLPPVNPAEE250 pKa = 4.79HH251 pKa = 6.51GNSVPALLEE260 pKa = 3.84HH261 pKa = 6.24VRR263 pKa = 11.84NLMIEE268 pKa = 4.11EE269 pKa = 4.25KK270 pKa = 10.79ARR272 pKa = 11.84LEE274 pKa = 3.94AALAARR280 pKa = 11.84RR281 pKa = 3.75
MM1 pKa = 7.64PWHH4 pKa = 6.72EE5 pKa = 4.57PVPHH9 pKa = 7.3RR10 pKa = 11.84PRR12 pKa = 11.84QRR14 pKa = 11.84AAPAFSLPGKK24 pKa = 9.16RR25 pKa = 11.84ACGTLRR31 pKa = 11.84PTIRR35 pKa = 11.84LLFSILFWSFFALSSLVFYY54 pKa = 11.03LGALVLWALTLPFDD68 pKa = 4.05RR69 pKa = 11.84NGRR72 pKa = 11.84ILHH75 pKa = 6.84LYY77 pKa = 8.69SCFWAQLYY85 pKa = 10.31IYY87 pKa = 9.82VNPLWSSRR95 pKa = 11.84VEE97 pKa = 4.07GRR99 pKa = 11.84EE100 pKa = 3.69NLPWRR105 pKa = 11.84GAAVLVANHH114 pKa = 6.58EE115 pKa = 4.46SLGDD119 pKa = 3.57ILVLFGLYY127 pKa = 10.06RR128 pKa = 11.84PFKK131 pKa = 9.35WVSKK135 pKa = 9.85AANFRR140 pKa = 11.84LPFIGWNMTLNRR152 pKa = 11.84YY153 pKa = 8.95VPLVRR158 pKa = 11.84GDD160 pKa = 3.64RR161 pKa = 11.84EE162 pKa = 4.35SVGRR166 pKa = 11.84MMVEE170 pKa = 3.85CEE172 pKa = 3.36KK173 pKa = 10.43WLLRR177 pKa = 11.84GVPILMFPEE186 pKa = 5.23GTRR189 pKa = 11.84SPDD192 pKa = 3.21GNIKK196 pKa = 10.35AFKK199 pKa = 10.66DD200 pKa = 3.68GAFQLAVKK208 pKa = 8.0LQCPVIPVVLTGTARR223 pKa = 11.84TLPKK227 pKa = 10.29HH228 pKa = 5.06GLKK231 pKa = 10.56LQTTARR237 pKa = 11.84CRR239 pKa = 11.84VRR241 pKa = 11.84VLPPVNPAEE250 pKa = 4.79HH251 pKa = 6.51GNSVPALLEE260 pKa = 3.84HH261 pKa = 6.24VRR263 pKa = 11.84NLMIEE268 pKa = 4.11EE269 pKa = 4.25KK270 pKa = 10.79ARR272 pKa = 11.84LEE274 pKa = 3.94AALAARR280 pKa = 11.84RR281 pKa = 3.75
Molecular weight: 31.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3100399 |
38 |
6261 |
373.2 |
40.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.601 ± 0.043 | 0.993 ± 0.012 |
4.7 ± 0.021 | 6.522 ± 0.034 |
3.408 ± 0.014 | 8.706 ± 0.03 |
2.164 ± 0.013 | 3.206 ± 0.018 |
3.055 ± 0.025 | 11.105 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.916 ± 0.011 | 2.324 ± 0.022 |
6.249 ± 0.025 | 3.753 ± 0.016 |
7.664 ± 0.035 | 6.013 ± 0.024 |
5.408 ± 0.04 | 7.633 ± 0.023 |
1.405 ± 0.012 | 2.175 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
