
Hubei unio douglasiae virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
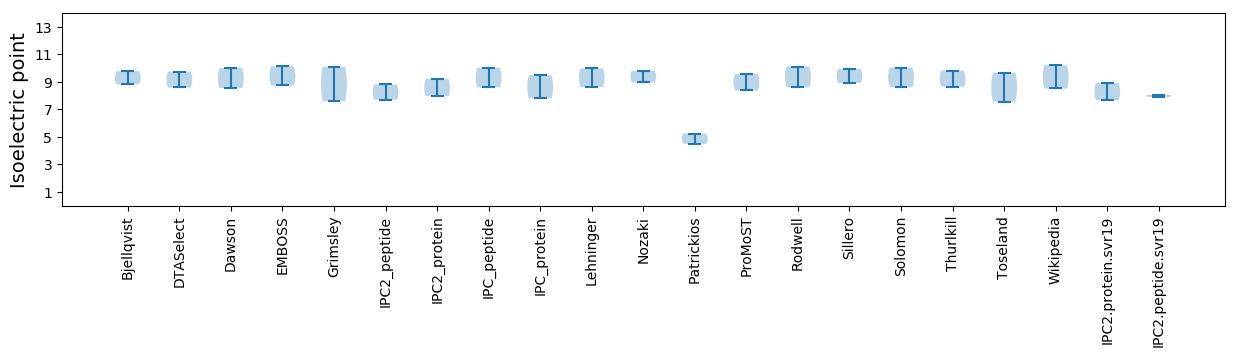
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGI3|A0A1L3KGI3_9VIRU RNA replicase OS=Hubei unio douglasiae virus 3 OX=1923323 PE=3 SV=1
MM1 pKa = 7.39AWWALAYY8 pKa = 9.6PLTQCMLGLWTSIALWQWVAFTRR31 pKa = 11.84APHH34 pKa = 6.65AYY36 pKa = 8.81AVAWGWLPEE45 pKa = 4.2CQSTGVWPWSRR56 pKa = 11.84DD57 pKa = 3.39TMLDD61 pKa = 3.2AAHH64 pKa = 5.75RR65 pKa = 11.84QVWRR69 pKa = 11.84FARR72 pKa = 11.84ARR74 pKa = 11.84GLCVEE79 pKa = 5.01PQHH82 pKa = 5.96SWWWLVIPPLCLILGYY98 pKa = 9.18TGARR102 pKa = 11.84CVRR105 pKa = 11.84YY106 pKa = 9.5GVLAVWFEE114 pKa = 4.52GVRR117 pKa = 11.84TLHH120 pKa = 6.6PKK122 pKa = 10.54ADD124 pKa = 3.93TKK126 pKa = 11.04RR127 pKa = 11.84KK128 pKa = 9.79LSGVKK133 pKa = 10.07CPPRR137 pKa = 11.84VATNPANPHH146 pKa = 5.67AAGAAIRR153 pKa = 11.84LQVHH157 pKa = 6.16KK158 pKa = 9.86FVHH161 pKa = 6.89AICAAWDD168 pKa = 3.44TGATVVRR175 pKa = 11.84YY176 pKa = 9.4DD177 pKa = 3.37LSYY180 pKa = 11.43AKK182 pKa = 9.74RR183 pKa = 11.84DD184 pKa = 3.66EE185 pKa = 4.25MFGTSVLGRR194 pKa = 11.84RR195 pKa = 11.84WLRR198 pKa = 11.84SAQDD202 pKa = 3.18LSQPVRR208 pKa = 11.84DD209 pKa = 4.24DD210 pKa = 3.46PRR212 pKa = 11.84PIGVPVSAIDD222 pKa = 3.14TAYY225 pKa = 11.13YY226 pKa = 9.73LRR228 pKa = 11.84PYY230 pKa = 10.62DD231 pKa = 3.38WDD233 pKa = 3.82EE234 pKa = 4.2FCGEE238 pKa = 4.59LIVFNAQLPTAIAWKK253 pKa = 9.95EE254 pKa = 4.04SWTTAYY260 pKa = 10.64FDD262 pKa = 3.88AQSVLHH268 pKa = 5.71EE269 pKa = 4.35QIAGGKK275 pKa = 7.72TYY277 pKa = 7.52THH279 pKa = 7.42PLPEE283 pKa = 5.31PGLHH287 pKa = 6.18EE288 pKa = 5.17LFLHH292 pKa = 5.48TRR294 pKa = 11.84GWFWNQRR301 pKa = 11.84VVCYY305 pKa = 8.67EE306 pKa = 3.74QTVVRR311 pKa = 11.84VPDD314 pKa = 3.26SRR316 pKa = 11.84QAIFIWTPQYY326 pKa = 10.94TITAPWWAFCAMYY339 pKa = 10.62RR340 pKa = 11.84FMTGSVPPRR349 pKa = 11.84PRR351 pKa = 11.84NWEE354 pKa = 3.46GAVRR358 pKa = 11.84VEE360 pKa = 4.74SSAGDD365 pKa = 3.26RR366 pKa = 11.84KK367 pKa = 10.5VSVVQMVTPDD377 pKa = 3.44GKK379 pKa = 10.36ISARR383 pKa = 11.84LEE385 pKa = 3.9STIDD389 pKa = 3.22GRR391 pKa = 11.84AFEE394 pKa = 4.52LDD396 pKa = 3.03RR397 pKa = 11.84TFVEE401 pKa = 5.26TVALRR406 pKa = 11.84GAVHH410 pKa = 6.78GSPVSIHH417 pKa = 6.66DD418 pKa = 3.68TSSMANRR425 pKa = 11.84MEE427 pKa = 4.47GVSTDD432 pKa = 3.56PAAVTMSAVVAGAGVLPVTTTPVGYY457 pKa = 10.54VPGAGVEE464 pKa = 4.26TAGARR469 pKa = 11.84VPTKK473 pKa = 10.58EE474 pKa = 3.84EE475 pKa = 4.59GIAGLVNPPASVAAHH490 pKa = 6.11TPAAVAAVLGRR501 pKa = 11.84VEE503 pKa = 4.09QLQVAVPRR511 pKa = 11.84TISPALKK518 pKa = 10.12SEE520 pKa = 3.89LMAAARR526 pKa = 11.84YY527 pKa = 9.43LGGLIGPMDD536 pKa = 4.28PKK538 pKa = 10.77SRR540 pKa = 11.84EE541 pKa = 3.85FVLEE545 pKa = 4.13SMTKK549 pKa = 8.42PAQRR553 pKa = 11.84ARR555 pKa = 11.84NAKK558 pKa = 10.04LPQQDD563 pKa = 3.73VAVMPDD569 pKa = 3.7AKK571 pKa = 10.28PSTMQKK577 pKa = 8.99TEE579 pKa = 4.09ATPKK583 pKa = 9.81PNPRR587 pKa = 11.84VINIVEE593 pKa = 3.83TDD595 pKa = 3.72FNVEE599 pKa = 3.68YY600 pKa = 11.28GRR602 pKa = 11.84FTKK605 pKa = 10.61AFYY608 pKa = 10.71AGLKK612 pKa = 8.77KK613 pKa = 10.76LPMCGSGVDD622 pKa = 3.81ATALAAKK629 pKa = 9.96IADD632 pKa = 3.78STVAGLLDD640 pKa = 4.61SIDD643 pKa = 3.72IADD646 pKa = 3.88MDD648 pKa = 4.34ASQGTTARR656 pKa = 11.84VDD658 pKa = 2.9IYY660 pKa = 10.5EE661 pKa = 4.07EE662 pKa = 3.98VMRR665 pKa = 11.84TVFAAHH671 pKa = 5.74LQEE674 pKa = 5.67LEE676 pKa = 4.15PLLLAARR683 pKa = 11.84TGKK686 pKa = 8.4LTYY689 pKa = 9.99RR690 pKa = 11.84SRR692 pKa = 11.84VDD694 pKa = 3.48DD695 pKa = 4.19AEE697 pKa = 4.42KK698 pKa = 10.06MVAVVSDD705 pKa = 4.51PNVSGCSDD713 pKa = 3.43TTNVNTIIMIVVALVAAQRR732 pKa = 11.84TDD734 pKa = 2.72GGYY737 pKa = 7.79DD738 pKa = 3.16THH740 pKa = 7.02PCGLFSGDD748 pKa = 4.35DD749 pKa = 3.46GLVPKK754 pKa = 10.37RR755 pKa = 11.84IRR757 pKa = 11.84SAFDD761 pKa = 2.92AVMAEE766 pKa = 4.04LGFTVKK772 pKa = 10.5VEE774 pKa = 4.32DD775 pKa = 3.96LASPEE780 pKa = 3.91AVKK783 pKa = 10.33FLNQIFVSPSTTRR796 pKa = 11.84TSTPSMHH803 pKa = 7.09RR804 pKa = 11.84LLSKK808 pKa = 10.74LCVITTANAGLLEE821 pKa = 5.1KK822 pKa = 10.76IMGLNISYY830 pKa = 9.29PRR832 pKa = 11.84NPIVQAVTRR841 pKa = 11.84AYY843 pKa = 9.12TNAYY847 pKa = 8.59GLAGADD853 pKa = 3.62MQRR856 pKa = 11.84EE857 pKa = 4.02RR858 pKa = 11.84ARR860 pKa = 11.84LKK862 pKa = 10.5KK863 pKa = 10.47RR864 pKa = 11.84PTYY867 pKa = 10.33EE868 pKa = 3.62LAAKK872 pKa = 10.07QEE874 pKa = 4.34AGAFLYY880 pKa = 10.47DD881 pKa = 3.98SKK883 pKa = 11.82DD884 pKa = 3.15EE885 pKa = 4.16AAVYY889 pKa = 10.49RR890 pKa = 11.84EE891 pKa = 4.01IAHH894 pKa = 6.43EE895 pKa = 4.31AGKK898 pKa = 8.94DD899 pKa = 3.8TAWVRR904 pKa = 11.84AWVKK908 pKa = 10.21EE909 pKa = 3.93LGKK912 pKa = 10.59VRR914 pKa = 11.84TQDD917 pKa = 3.74DD918 pKa = 3.94LLKK921 pKa = 11.11VGFLRR926 pKa = 11.84DD927 pKa = 3.9DD928 pKa = 3.86GSEE931 pKa = 4.0VDD933 pKa = 3.8GKK935 pKa = 11.26YY936 pKa = 10.35PITRR940 pKa = 11.84VAYY943 pKa = 8.45PAVPGKK949 pKa = 10.27GKK951 pKa = 10.63RR952 pKa = 11.84PGSSS956 pKa = 2.67
MM1 pKa = 7.39AWWALAYY8 pKa = 9.6PLTQCMLGLWTSIALWQWVAFTRR31 pKa = 11.84APHH34 pKa = 6.65AYY36 pKa = 8.81AVAWGWLPEE45 pKa = 4.2CQSTGVWPWSRR56 pKa = 11.84DD57 pKa = 3.39TMLDD61 pKa = 3.2AAHH64 pKa = 5.75RR65 pKa = 11.84QVWRR69 pKa = 11.84FARR72 pKa = 11.84ARR74 pKa = 11.84GLCVEE79 pKa = 5.01PQHH82 pKa = 5.96SWWWLVIPPLCLILGYY98 pKa = 9.18TGARR102 pKa = 11.84CVRR105 pKa = 11.84YY106 pKa = 9.5GVLAVWFEE114 pKa = 4.52GVRR117 pKa = 11.84TLHH120 pKa = 6.6PKK122 pKa = 10.54ADD124 pKa = 3.93TKK126 pKa = 11.04RR127 pKa = 11.84KK128 pKa = 9.79LSGVKK133 pKa = 10.07CPPRR137 pKa = 11.84VATNPANPHH146 pKa = 5.67AAGAAIRR153 pKa = 11.84LQVHH157 pKa = 6.16KK158 pKa = 9.86FVHH161 pKa = 6.89AICAAWDD168 pKa = 3.44TGATVVRR175 pKa = 11.84YY176 pKa = 9.4DD177 pKa = 3.37LSYY180 pKa = 11.43AKK182 pKa = 9.74RR183 pKa = 11.84DD184 pKa = 3.66EE185 pKa = 4.25MFGTSVLGRR194 pKa = 11.84RR195 pKa = 11.84WLRR198 pKa = 11.84SAQDD202 pKa = 3.18LSQPVRR208 pKa = 11.84DD209 pKa = 4.24DD210 pKa = 3.46PRR212 pKa = 11.84PIGVPVSAIDD222 pKa = 3.14TAYY225 pKa = 11.13YY226 pKa = 9.73LRR228 pKa = 11.84PYY230 pKa = 10.62DD231 pKa = 3.38WDD233 pKa = 3.82EE234 pKa = 4.2FCGEE238 pKa = 4.59LIVFNAQLPTAIAWKK253 pKa = 9.95EE254 pKa = 4.04SWTTAYY260 pKa = 10.64FDD262 pKa = 3.88AQSVLHH268 pKa = 5.71EE269 pKa = 4.35QIAGGKK275 pKa = 7.72TYY277 pKa = 7.52THH279 pKa = 7.42PLPEE283 pKa = 5.31PGLHH287 pKa = 6.18EE288 pKa = 5.17LFLHH292 pKa = 5.48TRR294 pKa = 11.84GWFWNQRR301 pKa = 11.84VVCYY305 pKa = 8.67EE306 pKa = 3.74QTVVRR311 pKa = 11.84VPDD314 pKa = 3.26SRR316 pKa = 11.84QAIFIWTPQYY326 pKa = 10.94TITAPWWAFCAMYY339 pKa = 10.62RR340 pKa = 11.84FMTGSVPPRR349 pKa = 11.84PRR351 pKa = 11.84NWEE354 pKa = 3.46GAVRR358 pKa = 11.84VEE360 pKa = 4.74SSAGDD365 pKa = 3.26RR366 pKa = 11.84KK367 pKa = 10.5VSVVQMVTPDD377 pKa = 3.44GKK379 pKa = 10.36ISARR383 pKa = 11.84LEE385 pKa = 3.9STIDD389 pKa = 3.22GRR391 pKa = 11.84AFEE394 pKa = 4.52LDD396 pKa = 3.03RR397 pKa = 11.84TFVEE401 pKa = 5.26TVALRR406 pKa = 11.84GAVHH410 pKa = 6.78GSPVSIHH417 pKa = 6.66DD418 pKa = 3.68TSSMANRR425 pKa = 11.84MEE427 pKa = 4.47GVSTDD432 pKa = 3.56PAAVTMSAVVAGAGVLPVTTTPVGYY457 pKa = 10.54VPGAGVEE464 pKa = 4.26TAGARR469 pKa = 11.84VPTKK473 pKa = 10.58EE474 pKa = 3.84EE475 pKa = 4.59GIAGLVNPPASVAAHH490 pKa = 6.11TPAAVAAVLGRR501 pKa = 11.84VEE503 pKa = 4.09QLQVAVPRR511 pKa = 11.84TISPALKK518 pKa = 10.12SEE520 pKa = 3.89LMAAARR526 pKa = 11.84YY527 pKa = 9.43LGGLIGPMDD536 pKa = 4.28PKK538 pKa = 10.77SRR540 pKa = 11.84EE541 pKa = 3.85FVLEE545 pKa = 4.13SMTKK549 pKa = 8.42PAQRR553 pKa = 11.84ARR555 pKa = 11.84NAKK558 pKa = 10.04LPQQDD563 pKa = 3.73VAVMPDD569 pKa = 3.7AKK571 pKa = 10.28PSTMQKK577 pKa = 8.99TEE579 pKa = 4.09ATPKK583 pKa = 9.81PNPRR587 pKa = 11.84VINIVEE593 pKa = 3.83TDD595 pKa = 3.72FNVEE599 pKa = 3.68YY600 pKa = 11.28GRR602 pKa = 11.84FTKK605 pKa = 10.61AFYY608 pKa = 10.71AGLKK612 pKa = 8.77KK613 pKa = 10.76LPMCGSGVDD622 pKa = 3.81ATALAAKK629 pKa = 9.96IADD632 pKa = 3.78STVAGLLDD640 pKa = 4.61SIDD643 pKa = 3.72IADD646 pKa = 3.88MDD648 pKa = 4.34ASQGTTARR656 pKa = 11.84VDD658 pKa = 2.9IYY660 pKa = 10.5EE661 pKa = 4.07EE662 pKa = 3.98VMRR665 pKa = 11.84TVFAAHH671 pKa = 5.74LQEE674 pKa = 5.67LEE676 pKa = 4.15PLLLAARR683 pKa = 11.84TGKK686 pKa = 8.4LTYY689 pKa = 9.99RR690 pKa = 11.84SRR692 pKa = 11.84VDD694 pKa = 3.48DD695 pKa = 4.19AEE697 pKa = 4.42KK698 pKa = 10.06MVAVVSDD705 pKa = 4.51PNVSGCSDD713 pKa = 3.43TTNVNTIIMIVVALVAAQRR732 pKa = 11.84TDD734 pKa = 2.72GGYY737 pKa = 7.79DD738 pKa = 3.16THH740 pKa = 7.02PCGLFSGDD748 pKa = 4.35DD749 pKa = 3.46GLVPKK754 pKa = 10.37RR755 pKa = 11.84IRR757 pKa = 11.84SAFDD761 pKa = 2.92AVMAEE766 pKa = 4.04LGFTVKK772 pKa = 10.5VEE774 pKa = 4.32DD775 pKa = 3.96LASPEE780 pKa = 3.91AVKK783 pKa = 10.33FLNQIFVSPSTTRR796 pKa = 11.84TSTPSMHH803 pKa = 7.09RR804 pKa = 11.84LLSKK808 pKa = 10.74LCVITTANAGLLEE821 pKa = 5.1KK822 pKa = 10.76IMGLNISYY830 pKa = 9.29PRR832 pKa = 11.84NPIVQAVTRR841 pKa = 11.84AYY843 pKa = 9.12TNAYY847 pKa = 8.59GLAGADD853 pKa = 3.62MQRR856 pKa = 11.84EE857 pKa = 4.02RR858 pKa = 11.84ARR860 pKa = 11.84LKK862 pKa = 10.5KK863 pKa = 10.47RR864 pKa = 11.84PTYY867 pKa = 10.33EE868 pKa = 3.62LAAKK872 pKa = 10.07QEE874 pKa = 4.34AGAFLYY880 pKa = 10.47DD881 pKa = 3.98SKK883 pKa = 11.82DD884 pKa = 3.15EE885 pKa = 4.16AAVYY889 pKa = 10.49RR890 pKa = 11.84EE891 pKa = 4.01IAHH894 pKa = 6.43EE895 pKa = 4.31AGKK898 pKa = 8.94DD899 pKa = 3.8TAWVRR904 pKa = 11.84AWVKK908 pKa = 10.21EE909 pKa = 3.93LGKK912 pKa = 10.59VRR914 pKa = 11.84TQDD917 pKa = 3.74DD918 pKa = 3.94LLKK921 pKa = 11.11VGFLRR926 pKa = 11.84DD927 pKa = 3.9DD928 pKa = 3.86GSEE931 pKa = 4.0VDD933 pKa = 3.8GKK935 pKa = 11.26YY936 pKa = 10.35PITRR940 pKa = 11.84VAYY943 pKa = 8.45PAVPGKK949 pKa = 10.27GKK951 pKa = 10.63RR952 pKa = 11.84PGSSS956 pKa = 2.67
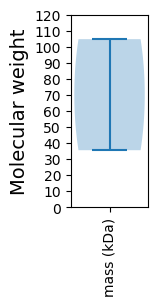
Molecular weight: 104.73 kDa
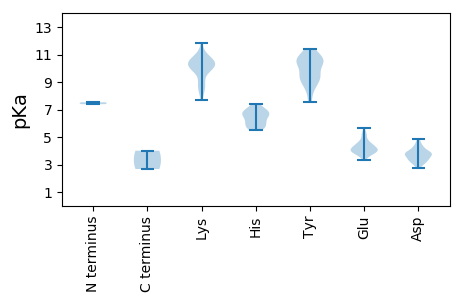
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGI3|A0A1L3KGI3_9VIRU RNA replicase OS=Hubei unio douglasiae virus 3 OX=1923323 PE=3 SV=1
MM1 pKa = 7.53PPKK4 pKa = 10.2AAEE7 pKa = 3.78QRR9 pKa = 11.84IVVAVAKK16 pKa = 10.31GVSQGAANAAKK27 pKa = 10.0QSKK30 pKa = 9.02ARR32 pKa = 11.84KK33 pKa = 8.4GGKK36 pKa = 8.28AQRR39 pKa = 11.84QRR41 pKa = 11.84MSIVPRR47 pKa = 11.84SVRR50 pKa = 11.84QLDD53 pKa = 3.95PPAMAWLKK61 pKa = 9.74MLNDD65 pKa = 3.64PCYY68 pKa = 10.78GRR70 pKa = 11.84LSHH73 pKa = 6.72PVYY76 pKa = 10.36PGADD80 pKa = 2.97GGYY83 pKa = 9.48LSRR86 pKa = 11.84FEE88 pKa = 4.76SEE90 pKa = 3.93ATFVAGVGTTAGIIGFVPGVLPFAVVGSGVVSDD123 pKa = 3.74ITASALADD131 pKa = 4.12LSAASAPGYY140 pKa = 9.18TYY142 pKa = 11.12LRR144 pKa = 11.84GVANSVRR151 pKa = 11.84CVSACMQVSWPGSEE165 pKa = 4.64LNRR168 pKa = 11.84QGFVTLGQSTGAVLAEE184 pKa = 3.8AAAAFGNTPVTPASLRR200 pKa = 11.84PLCHH204 pKa = 6.53LRR206 pKa = 11.84TRR208 pKa = 11.84MPEE211 pKa = 3.71TVAEE215 pKa = 4.56VKK217 pKa = 9.12WRR219 pKa = 11.84PTLADD224 pKa = 4.08AQWHH228 pKa = 6.97DD229 pKa = 4.05PLVAPSYY236 pKa = 10.57GRR238 pKa = 11.84VNEE241 pKa = 4.26AGALLATFGNLPLDD255 pKa = 3.75VNGNGIGVRR264 pKa = 11.84VRR266 pKa = 11.84FIVTYY271 pKa = 9.1EE272 pKa = 4.18WIPRR276 pKa = 11.84GAQGLTSGFDD286 pKa = 3.24DD287 pKa = 4.33RR288 pKa = 11.84ARR290 pKa = 11.84SSNTLDD296 pKa = 4.7DD297 pKa = 4.85VINTLDD303 pKa = 3.52RR304 pKa = 11.84NGPEE308 pKa = 3.36WAYY311 pKa = 9.83TIGRR315 pKa = 11.84ATSVLGMMGSAYY327 pKa = 10.43VAGRR331 pKa = 11.84RR332 pKa = 11.84NRR334 pKa = 11.84LMGG337 pKa = 4.01
MM1 pKa = 7.53PPKK4 pKa = 10.2AAEE7 pKa = 3.78QRR9 pKa = 11.84IVVAVAKK16 pKa = 10.31GVSQGAANAAKK27 pKa = 10.0QSKK30 pKa = 9.02ARR32 pKa = 11.84KK33 pKa = 8.4GGKK36 pKa = 8.28AQRR39 pKa = 11.84QRR41 pKa = 11.84MSIVPRR47 pKa = 11.84SVRR50 pKa = 11.84QLDD53 pKa = 3.95PPAMAWLKK61 pKa = 9.74MLNDD65 pKa = 3.64PCYY68 pKa = 10.78GRR70 pKa = 11.84LSHH73 pKa = 6.72PVYY76 pKa = 10.36PGADD80 pKa = 2.97GGYY83 pKa = 9.48LSRR86 pKa = 11.84FEE88 pKa = 4.76SEE90 pKa = 3.93ATFVAGVGTTAGIIGFVPGVLPFAVVGSGVVSDD123 pKa = 3.74ITASALADD131 pKa = 4.12LSAASAPGYY140 pKa = 9.18TYY142 pKa = 11.12LRR144 pKa = 11.84GVANSVRR151 pKa = 11.84CVSACMQVSWPGSEE165 pKa = 4.64LNRR168 pKa = 11.84QGFVTLGQSTGAVLAEE184 pKa = 3.8AAAAFGNTPVTPASLRR200 pKa = 11.84PLCHH204 pKa = 6.53LRR206 pKa = 11.84TRR208 pKa = 11.84MPEE211 pKa = 3.71TVAEE215 pKa = 4.56VKK217 pKa = 9.12WRR219 pKa = 11.84PTLADD224 pKa = 4.08AQWHH228 pKa = 6.97DD229 pKa = 4.05PLVAPSYY236 pKa = 10.57GRR238 pKa = 11.84VNEE241 pKa = 4.26AGALLATFGNLPLDD255 pKa = 3.75VNGNGIGVRR264 pKa = 11.84VRR266 pKa = 11.84FIVTYY271 pKa = 9.1EE272 pKa = 4.18WIPRR276 pKa = 11.84GAQGLTSGFDD286 pKa = 3.24DD287 pKa = 4.33RR288 pKa = 11.84ARR290 pKa = 11.84SSNTLDD296 pKa = 4.7DD297 pKa = 4.85VINTLDD303 pKa = 3.52RR304 pKa = 11.84NGPEE308 pKa = 3.36WAYY311 pKa = 9.83TIGRR315 pKa = 11.84ATSVLGMMGSAYY327 pKa = 10.43VAGRR331 pKa = 11.84RR332 pKa = 11.84NRR334 pKa = 11.84LMGG337 pKa = 4.01
Molecular weight: 35.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1293 |
337 |
956 |
646.5 |
70.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.684 ± 0.283 | 1.392 ± 0.087 |
4.872 ± 0.429 | 4.176 ± 0.512 |
2.784 ± 0.048 | 7.966 ± 1.275 |
1.624 ± 0.311 | 3.558 ± 0.25 |
3.712 ± 0.566 | 7.579 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.05 | 2.475 ± 0.585 |
6.574 ± 0.019 | 3.171 ± 0.039 |
7.038 ± 0.287 | 5.878 ± 0.526 |
6.806 ± 0.494 | 9.667 ± 0.053 |
2.552 ± 0.327 | 2.939 ± 0.114 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
