
African elephant polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Loxodonta africana polyomavirus 1
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
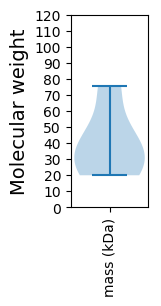
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|T2FFJ0|T2FFJ0_9POLY Minor capsid protein OS=African elephant polyomavirus 1 OX=1399914 GN=AelPyV_gp2 PE=3 SV=1
MM1 pKa = 7.67APQRR5 pKa = 11.84KK6 pKa = 9.29GGGRR10 pKa = 11.84PCVPSKK16 pKa = 10.88RR17 pKa = 11.84PLRR20 pKa = 11.84KK21 pKa = 9.26PEE23 pKa = 3.85NVPRR27 pKa = 11.84ILVRR31 pKa = 11.84GGVQVLEE38 pKa = 4.43VRR40 pKa = 11.84SGPDD44 pKa = 3.05SIVDD48 pKa = 3.48IEE50 pKa = 5.04CFLQPRR56 pKa = 11.84MGEE59 pKa = 4.13PEE61 pKa = 3.99GDD63 pKa = 3.39DD64 pKa = 3.49YY65 pKa = 11.96YY66 pKa = 11.51GYY68 pKa = 10.66SEE70 pKa = 5.98LITVGTEE77 pKa = 3.54KK78 pKa = 10.25STDD81 pKa = 3.36TPPFKK86 pKa = 10.46QLPCYY91 pKa = 10.29SAAQVQLPNLNPDD104 pKa = 4.24LTCEE108 pKa = 3.95TLLMWEE114 pKa = 4.21AVSTKK119 pKa = 10.34TEE121 pKa = 3.82VLGISSLVNVHH132 pKa = 5.5SQSKK136 pKa = 9.0RR137 pKa = 11.84VEE139 pKa = 4.14PEE141 pKa = 4.18GIGYY145 pKa = 8.45PIAGINYY152 pKa = 8.5HH153 pKa = 6.19FFAVGGQPLDD163 pKa = 3.73LQALVSNFSATYY175 pKa = 7.56PAEE178 pKa = 3.98RR179 pKa = 11.84KK180 pKa = 10.05VPVKK184 pKa = 10.78DD185 pKa = 3.34QLHH188 pKa = 7.18PIAQVLDD195 pKa = 4.22PNLKK199 pKa = 7.71TTLDD203 pKa = 3.63EE204 pKa = 5.35DD205 pKa = 3.54GTYY208 pKa = 10.3PIEE211 pKa = 4.2VWGADD216 pKa = 3.31PSKK219 pKa = 11.25NDD221 pKa = 2.94NTRR224 pKa = 11.84YY225 pKa = 10.0FGTLTGGAQTPPVLSVTNTTTTILLDD251 pKa = 3.95EE252 pKa = 4.77YY253 pKa = 11.48GVGPLCKK260 pKa = 10.15GGRR263 pKa = 11.84LYY265 pKa = 10.93LAAADD270 pKa = 3.2ICGFSTEE277 pKa = 4.39STGKK281 pKa = 8.46MRR283 pKa = 11.84YY284 pKa = 9.24RR285 pKa = 11.84GLPRR289 pKa = 11.84FFKK292 pKa = 10.74VKK294 pKa = 9.57LRR296 pKa = 11.84KK297 pKa = 9.31RR298 pKa = 11.84AVKK301 pKa = 10.12NPYY304 pKa = 10.41AIGSLLNSFFSSFLPQVKK322 pKa = 8.75GQPMTGDD329 pKa = 3.0KK330 pKa = 11.05GQVEE334 pKa = 4.93EE335 pKa = 4.2VTIFQGTEE343 pKa = 3.89PLPADD348 pKa = 4.02PDD350 pKa = 4.46LIRR353 pKa = 11.84SVDD356 pKa = 3.36QFGQFLTKK364 pKa = 10.38EE365 pKa = 3.93PAQANVVV372 pKa = 3.43
MM1 pKa = 7.67APQRR5 pKa = 11.84KK6 pKa = 9.29GGGRR10 pKa = 11.84PCVPSKK16 pKa = 10.88RR17 pKa = 11.84PLRR20 pKa = 11.84KK21 pKa = 9.26PEE23 pKa = 3.85NVPRR27 pKa = 11.84ILVRR31 pKa = 11.84GGVQVLEE38 pKa = 4.43VRR40 pKa = 11.84SGPDD44 pKa = 3.05SIVDD48 pKa = 3.48IEE50 pKa = 5.04CFLQPRR56 pKa = 11.84MGEE59 pKa = 4.13PEE61 pKa = 3.99GDD63 pKa = 3.39DD64 pKa = 3.49YY65 pKa = 11.96YY66 pKa = 11.51GYY68 pKa = 10.66SEE70 pKa = 5.98LITVGTEE77 pKa = 3.54KK78 pKa = 10.25STDD81 pKa = 3.36TPPFKK86 pKa = 10.46QLPCYY91 pKa = 10.29SAAQVQLPNLNPDD104 pKa = 4.24LTCEE108 pKa = 3.95TLLMWEE114 pKa = 4.21AVSTKK119 pKa = 10.34TEE121 pKa = 3.82VLGISSLVNVHH132 pKa = 5.5SQSKK136 pKa = 9.0RR137 pKa = 11.84VEE139 pKa = 4.14PEE141 pKa = 4.18GIGYY145 pKa = 8.45PIAGINYY152 pKa = 8.5HH153 pKa = 6.19FFAVGGQPLDD163 pKa = 3.73LQALVSNFSATYY175 pKa = 7.56PAEE178 pKa = 3.98RR179 pKa = 11.84KK180 pKa = 10.05VPVKK184 pKa = 10.78DD185 pKa = 3.34QLHH188 pKa = 7.18PIAQVLDD195 pKa = 4.22PNLKK199 pKa = 7.71TTLDD203 pKa = 3.63EE204 pKa = 5.35DD205 pKa = 3.54GTYY208 pKa = 10.3PIEE211 pKa = 4.2VWGADD216 pKa = 3.31PSKK219 pKa = 11.25NDD221 pKa = 2.94NTRR224 pKa = 11.84YY225 pKa = 10.0FGTLTGGAQTPPVLSVTNTTTTILLDD251 pKa = 3.95EE252 pKa = 4.77YY253 pKa = 11.48GVGPLCKK260 pKa = 10.15GGRR263 pKa = 11.84LYY265 pKa = 10.93LAAADD270 pKa = 3.2ICGFSTEE277 pKa = 4.39STGKK281 pKa = 8.46MRR283 pKa = 11.84YY284 pKa = 9.24RR285 pKa = 11.84GLPRR289 pKa = 11.84FFKK292 pKa = 10.74VKK294 pKa = 9.57LRR296 pKa = 11.84KK297 pKa = 9.31RR298 pKa = 11.84AVKK301 pKa = 10.12NPYY304 pKa = 10.41AIGSLLNSFFSSFLPQVKK322 pKa = 8.75GQPMTGDD329 pKa = 3.0KK330 pKa = 11.05GQVEE334 pKa = 4.93EE335 pKa = 4.2VTIFQGTEE343 pKa = 3.89PLPADD348 pKa = 4.02PDD350 pKa = 4.46LIRR353 pKa = 11.84SVDD356 pKa = 3.36QFGQFLTKK364 pKa = 10.38EE365 pKa = 3.93PAQANVVV372 pKa = 3.43
Molecular weight: 40.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T2FFC5|T2FFC5_9POLY Major capsid protein VP1 OS=African elephant polyomavirus 1 OX=1399914 GN=AelPyV_gp1 PE=3 SV=1
MM1 pKa = 7.76ALQLWFPPVDD11 pKa = 3.49YY12 pKa = 10.53LFPGVTQFSRR22 pKa = 11.84FLRR25 pKa = 11.84YY26 pKa = 9.0IDD28 pKa = 3.49PTQWGPEE35 pKa = 4.1LFHH38 pKa = 7.14HH39 pKa = 6.48LTQAFFSTFRR49 pKa = 11.84DD50 pKa = 3.37ATRR53 pKa = 11.84RR54 pKa = 11.84QLTDD58 pKa = 2.99ASRR61 pKa = 11.84NIAEE65 pKa = 4.33RR66 pKa = 11.84TAKK69 pKa = 9.83TLVEE73 pKa = 4.21TLVDD77 pKa = 4.46LLTNATWAIAHH88 pKa = 6.75LPSDD92 pKa = 4.02VYY94 pKa = 11.21TFLDD98 pKa = 3.26NYY100 pKa = 10.78YY101 pKa = 10.89EE102 pKa = 4.22EE103 pKa = 5.58LPPLNPAQFSGIRR116 pKa = 11.84KK117 pKa = 9.35RR118 pKa = 11.84LGKK121 pKa = 10.16VNPYY125 pKa = 8.73EE126 pKa = 4.12QQDD129 pKa = 3.7VFANTVSAEE138 pKa = 3.99FVTKK142 pKa = 9.27VTAPGGAMQKK152 pKa = 9.93HH153 pKa = 5.51APDD156 pKa = 3.27WVLPLLLGLFDD167 pKa = 3.75EE168 pKa = 6.29DD169 pKa = 3.92YY170 pKa = 11.08RR171 pKa = 11.84QVTPRR176 pKa = 11.84DD177 pKa = 3.21GPAAKK182 pKa = 9.86RR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84TTLRR189 pKa = 11.84TIQKK193 pKa = 7.69TPPKK197 pKa = 8.96TGKK200 pKa = 10.13RR201 pKa = 11.84PPNTRR206 pKa = 11.84ARR208 pKa = 11.84RR209 pKa = 11.84SASSRR214 pKa = 11.84SKK216 pKa = 10.74KK217 pKa = 9.85RR218 pKa = 11.84AA219 pKa = 3.11
MM1 pKa = 7.76ALQLWFPPVDD11 pKa = 3.49YY12 pKa = 10.53LFPGVTQFSRR22 pKa = 11.84FLRR25 pKa = 11.84YY26 pKa = 9.0IDD28 pKa = 3.49PTQWGPEE35 pKa = 4.1LFHH38 pKa = 7.14HH39 pKa = 6.48LTQAFFSTFRR49 pKa = 11.84DD50 pKa = 3.37ATRR53 pKa = 11.84RR54 pKa = 11.84QLTDD58 pKa = 2.99ASRR61 pKa = 11.84NIAEE65 pKa = 4.33RR66 pKa = 11.84TAKK69 pKa = 9.83TLVEE73 pKa = 4.21TLVDD77 pKa = 4.46LLTNATWAIAHH88 pKa = 6.75LPSDD92 pKa = 4.02VYY94 pKa = 11.21TFLDD98 pKa = 3.26NYY100 pKa = 10.78YY101 pKa = 10.89EE102 pKa = 4.22EE103 pKa = 5.58LPPLNPAQFSGIRR116 pKa = 11.84KK117 pKa = 9.35RR118 pKa = 11.84LGKK121 pKa = 10.16VNPYY125 pKa = 8.73EE126 pKa = 4.12QQDD129 pKa = 3.7VFANTVSAEE138 pKa = 3.99FVTKK142 pKa = 9.27VTAPGGAMQKK152 pKa = 9.93HH153 pKa = 5.51APDD156 pKa = 3.27WVLPLLLGLFDD167 pKa = 3.75EE168 pKa = 6.29DD169 pKa = 3.92YY170 pKa = 11.08RR171 pKa = 11.84QVTPRR176 pKa = 11.84DD177 pKa = 3.21GPAAKK182 pKa = 9.86RR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84TTLRR189 pKa = 11.84TIQKK193 pKa = 7.69TPPKK197 pKa = 8.96TGKK200 pKa = 10.13RR201 pKa = 11.84PPNTRR206 pKa = 11.84ARR208 pKa = 11.84RR209 pKa = 11.84SASSRR214 pKa = 11.84SKK216 pKa = 10.74KK217 pKa = 9.85RR218 pKa = 11.84AA219 pKa = 3.11
Molecular weight: 25.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1750 |
171 |
658 |
350.0 |
39.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.686 ± 1.484 | 2.4 ± 0.905 |
5.657 ± 0.703 | 5.2 ± 0.241 |
5.314 ± 0.433 | 5.943 ± 0.692 |
2.114 ± 0.387 | 3.143 ± 0.431 |
5.886 ± 0.801 | 10.514 ± 0.426 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.4 ± 0.746 | 4.286 ± 0.545 |
6.4 ± 0.945 | 4.171 ± 0.31 |
6.057 ± 0.762 | 6.114 ± 0.276 |
7.314 ± 0.7 | 5.771 ± 0.672 |
1.371 ± 0.278 | 3.257 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
