
Hubei sobemo-like virus 40
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
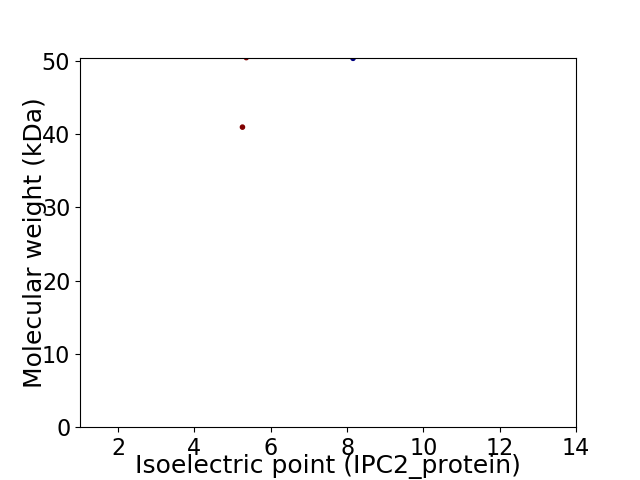
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEJ8|A0A1L3KEJ8_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 40 OX=1923228 PE=4 SV=1
MM1 pKa = 6.85TQAPTIGEE9 pKa = 4.02WLFKK13 pKa = 11.31GEE15 pKa = 4.83LEE17 pKa = 4.25PDD19 pKa = 3.53PTRR22 pKa = 11.84KK23 pKa = 9.41EE24 pKa = 4.22LLWLMVTDD32 pKa = 4.53VIEE35 pKa = 4.36GNYY38 pKa = 9.08EE39 pKa = 3.5HH40 pKa = 7.57LFRR43 pKa = 11.84VFVKK47 pKa = 10.57HH48 pKa = 6.04EE49 pKa = 3.7PHH51 pKa = 6.37TEE53 pKa = 3.62RR54 pKa = 11.84KK55 pKa = 9.68ALAKK59 pKa = 9.84RR60 pKa = 11.84WRR62 pKa = 11.84LIIASSLPVQIVWQMVFGNIEE83 pKa = 4.12DD84 pKa = 4.86ALLQQTGRR92 pKa = 11.84TPSAYY97 pKa = 10.1GLVYY101 pKa = 10.41GAGGWKK107 pKa = 10.04RR108 pKa = 11.84FRR110 pKa = 11.84DD111 pKa = 3.46MLVQRR116 pKa = 11.84KK117 pKa = 8.64VNLCVDD123 pKa = 3.56KK124 pKa = 11.38SGWDD128 pKa = 3.42MNSPGWVYY136 pKa = 9.38EE137 pKa = 3.92LCLEE141 pKa = 4.08LRR143 pKa = 11.84KK144 pKa = 9.84RR145 pKa = 11.84LCTDD149 pKa = 3.98LTPAAEE155 pKa = 4.07RR156 pKa = 11.84VMDD159 pKa = 3.75MLYY162 pKa = 10.47RR163 pKa = 11.84DD164 pKa = 4.77AYY166 pKa = 10.49QSSKK170 pKa = 11.13LIFGDD175 pKa = 4.02GTILEE180 pKa = 4.31QQFSGFMKK188 pKa = 10.24SGLFVTISDD197 pKa = 3.93NSFSQYY203 pKa = 10.13FLHH206 pKa = 7.01VLACLRR212 pKa = 11.84LRR214 pKa = 11.84IPLGTFYY221 pKa = 10.75ATGDD225 pKa = 3.67DD226 pKa = 4.7TIQSLPPNVEE236 pKa = 4.0EE237 pKa = 4.35YY238 pKa = 11.01LDD240 pKa = 3.67HH241 pKa = 7.71LEE243 pKa = 4.16YY244 pKa = 10.83AGCEE248 pKa = 3.86VKK250 pKa = 10.43EE251 pKa = 4.07YY252 pKa = 10.1MNGYY256 pKa = 9.37QFMGFEE262 pKa = 4.26LKK264 pKa = 10.44DD265 pKa = 3.77YY266 pKa = 10.17GPHH269 pKa = 6.96PIYY272 pKa = 10.39LGKK275 pKa = 10.13HH276 pKa = 5.02FWNLLHH282 pKa = 7.3QEE284 pKa = 4.37DD285 pKa = 4.79EE286 pKa = 4.37NLEE289 pKa = 4.06QTIDD293 pKa = 3.58SYY295 pKa = 12.0LLNYY299 pKa = 9.88CKK301 pKa = 10.31VDD303 pKa = 3.31EE304 pKa = 4.72AFDD307 pKa = 3.82YY308 pKa = 11.15LRR310 pKa = 11.84ALSIEE315 pKa = 4.29LGYY318 pKa = 11.0APRR321 pKa = 11.84SKK323 pKa = 9.9QWYY326 pKa = 8.5RR327 pKa = 11.84FLMDD331 pKa = 4.11NPLALEE337 pKa = 4.53GNWTRR342 pKa = 11.84PGFVDD347 pKa = 3.5PASRR351 pKa = 11.84LGG353 pKa = 3.41
MM1 pKa = 6.85TQAPTIGEE9 pKa = 4.02WLFKK13 pKa = 11.31GEE15 pKa = 4.83LEE17 pKa = 4.25PDD19 pKa = 3.53PTRR22 pKa = 11.84KK23 pKa = 9.41EE24 pKa = 4.22LLWLMVTDD32 pKa = 4.53VIEE35 pKa = 4.36GNYY38 pKa = 9.08EE39 pKa = 3.5HH40 pKa = 7.57LFRR43 pKa = 11.84VFVKK47 pKa = 10.57HH48 pKa = 6.04EE49 pKa = 3.7PHH51 pKa = 6.37TEE53 pKa = 3.62RR54 pKa = 11.84KK55 pKa = 9.68ALAKK59 pKa = 9.84RR60 pKa = 11.84WRR62 pKa = 11.84LIIASSLPVQIVWQMVFGNIEE83 pKa = 4.12DD84 pKa = 4.86ALLQQTGRR92 pKa = 11.84TPSAYY97 pKa = 10.1GLVYY101 pKa = 10.41GAGGWKK107 pKa = 10.04RR108 pKa = 11.84FRR110 pKa = 11.84DD111 pKa = 3.46MLVQRR116 pKa = 11.84KK117 pKa = 8.64VNLCVDD123 pKa = 3.56KK124 pKa = 11.38SGWDD128 pKa = 3.42MNSPGWVYY136 pKa = 9.38EE137 pKa = 3.92LCLEE141 pKa = 4.08LRR143 pKa = 11.84KK144 pKa = 9.84RR145 pKa = 11.84LCTDD149 pKa = 3.98LTPAAEE155 pKa = 4.07RR156 pKa = 11.84VMDD159 pKa = 3.75MLYY162 pKa = 10.47RR163 pKa = 11.84DD164 pKa = 4.77AYY166 pKa = 10.49QSSKK170 pKa = 11.13LIFGDD175 pKa = 4.02GTILEE180 pKa = 4.31QQFSGFMKK188 pKa = 10.24SGLFVTISDD197 pKa = 3.93NSFSQYY203 pKa = 10.13FLHH206 pKa = 7.01VLACLRR212 pKa = 11.84LRR214 pKa = 11.84IPLGTFYY221 pKa = 10.75ATGDD225 pKa = 3.67DD226 pKa = 4.7TIQSLPPNVEE236 pKa = 4.0EE237 pKa = 4.35YY238 pKa = 11.01LDD240 pKa = 3.67HH241 pKa = 7.71LEE243 pKa = 4.16YY244 pKa = 10.83AGCEE248 pKa = 3.86VKK250 pKa = 10.43EE251 pKa = 4.07YY252 pKa = 10.1MNGYY256 pKa = 9.37QFMGFEE262 pKa = 4.26LKK264 pKa = 10.44DD265 pKa = 3.77YY266 pKa = 10.17GPHH269 pKa = 6.96PIYY272 pKa = 10.39LGKK275 pKa = 10.13HH276 pKa = 5.02FWNLLHH282 pKa = 7.3QEE284 pKa = 4.37DD285 pKa = 4.79EE286 pKa = 4.37NLEE289 pKa = 4.06QTIDD293 pKa = 3.58SYY295 pKa = 12.0LLNYY299 pKa = 9.88CKK301 pKa = 10.31VDD303 pKa = 3.31EE304 pKa = 4.72AFDD307 pKa = 3.82YY308 pKa = 11.15LRR310 pKa = 11.84ALSIEE315 pKa = 4.29LGYY318 pKa = 11.0APRR321 pKa = 11.84SKK323 pKa = 9.9QWYY326 pKa = 8.5RR327 pKa = 11.84FLMDD331 pKa = 4.11NPLALEE337 pKa = 4.53GNWTRR342 pKa = 11.84PGFVDD347 pKa = 3.5PASRR351 pKa = 11.84LGG353 pKa = 3.41
Molecular weight: 40.97 kDa
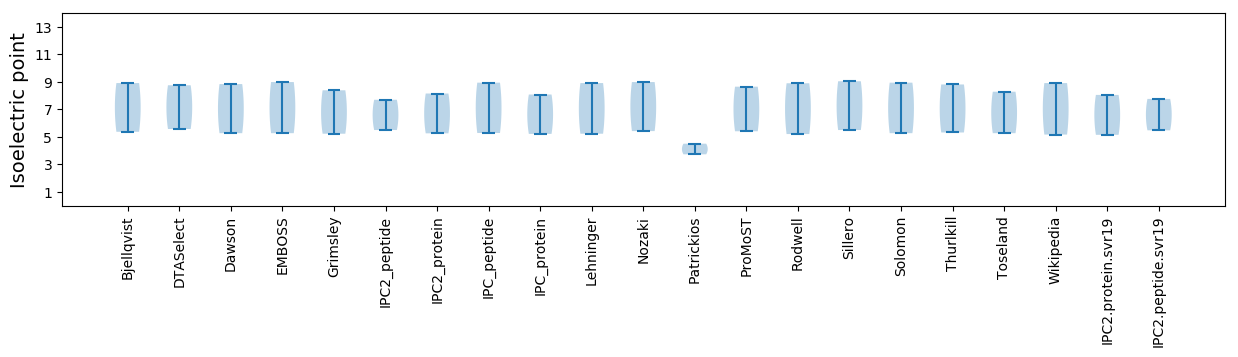
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEJ8|A0A1L3KEJ8_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 40 OX=1923228 PE=4 SV=1
MM1 pKa = 6.62YY2 pKa = 9.26TNLYY6 pKa = 9.1PHH8 pKa = 6.85FSAAKK13 pKa = 9.5SLAFQSGIEE22 pKa = 4.0KK23 pKa = 10.43SALVGKK29 pKa = 10.26AVTEE33 pKa = 4.26TVKK36 pKa = 10.92ARR38 pKa = 11.84VNLTRR43 pKa = 11.84LAVEE47 pKa = 4.42TVKK50 pKa = 10.77EE51 pKa = 4.16DD52 pKa = 3.88PLRR55 pKa = 11.84AALTVTSIVSAGWVLKK71 pKa = 10.43KK72 pKa = 10.37AVPFAYY78 pKa = 9.43RR79 pKa = 11.84KK80 pKa = 8.14YY81 pKa = 9.89RR82 pKa = 11.84EE83 pKa = 3.92WRR85 pKa = 11.84EE86 pKa = 3.98RR87 pKa = 11.84PWPVIKK93 pKa = 10.0EE94 pKa = 4.14APSYY98 pKa = 10.11EE99 pKa = 4.24PEE101 pKa = 4.13TAVEE105 pKa = 4.94GSVEE109 pKa = 4.35TNMSFARR116 pKa = 11.84CQGALCVAEE125 pKa = 4.24GQTWKK130 pKa = 9.36TVGGCFRR137 pKa = 11.84LTVGSKK143 pKa = 10.87DD144 pKa = 3.64LLLTAGHH151 pKa = 6.37NLVEE155 pKa = 4.32GEE157 pKa = 4.28RR158 pKa = 11.84YY159 pKa = 9.43AVSRR163 pKa = 11.84MGKK166 pKa = 9.16IKK168 pKa = 10.43EE169 pKa = 4.38LGDD172 pKa = 3.06VWSAKK177 pKa = 10.44DD178 pKa = 3.19ADD180 pKa = 4.24QIYY183 pKa = 10.8SPVTDD188 pKa = 3.91IVAIPLPDD196 pKa = 3.39SFFSGLSMRR205 pKa = 11.84SATTIALPSSTNAQITGALGKK226 pKa = 10.76GSVGKK231 pKa = 8.94LHH233 pKa = 7.22PLGQFGRR240 pKa = 11.84VEE242 pKa = 4.2YY243 pKa = 10.79AGTTAGGYY251 pKa = 9.21SGCPYY256 pKa = 10.26IVGEE260 pKa = 4.24RR261 pKa = 11.84VAAIHH266 pKa = 5.97TNGGRR271 pKa = 11.84KK272 pKa = 9.27NEE274 pKa = 3.82GWEE277 pKa = 3.85IKK279 pKa = 10.5YY280 pKa = 9.76VEE282 pKa = 4.48CLLYY286 pKa = 11.07AHH288 pKa = 6.5FVVSNEE294 pKa = 4.08STVGSEE300 pKa = 3.83DD301 pKa = 3.18AARR304 pKa = 11.84RR305 pKa = 11.84AAQEE309 pKa = 3.87EE310 pKa = 4.59HH311 pKa = 7.48DD312 pKa = 3.99IAPYY316 pKa = 10.8DD317 pKa = 4.26DD318 pKa = 4.89DD319 pKa = 5.97SIIIRR324 pKa = 11.84SRR326 pKa = 11.84KK327 pKa = 6.34TGKK330 pKa = 10.24YY331 pKa = 7.82YY332 pKa = 9.6RR333 pKa = 11.84TRR335 pKa = 11.84EE336 pKa = 3.96TVYY339 pKa = 10.83EE340 pKa = 3.89EE341 pKa = 4.39LMRR344 pKa = 11.84AKK346 pKa = 10.07QSYY349 pKa = 10.38ANNPDD354 pKa = 2.7RR355 pKa = 11.84WADD358 pKa = 3.57QVEE361 pKa = 4.27VDD363 pKa = 5.41LLQQEE368 pKa = 4.68LDD370 pKa = 3.6SYY372 pKa = 10.83MPEE375 pKa = 4.32CASEE379 pKa = 4.28AGNAKK384 pKa = 9.0TPAQAGVGQLDD395 pKa = 3.39GDD397 pKa = 3.86MSVKK401 pKa = 10.3QFLSLLRR408 pKa = 11.84EE409 pKa = 3.87NRR411 pKa = 11.84ALSMSEE417 pKa = 4.07SSEE420 pKa = 3.75QSQLSRR426 pKa = 11.84RR427 pKa = 11.84QRR429 pKa = 11.84RR430 pKa = 11.84QAYY433 pKa = 8.31FGRR436 pKa = 11.84RR437 pKa = 11.84MGLEE441 pKa = 3.48RR442 pKa = 11.84QANIRR447 pKa = 11.84RR448 pKa = 11.84RR449 pKa = 11.84SPTPARR455 pKa = 11.84KK456 pKa = 9.25
MM1 pKa = 6.62YY2 pKa = 9.26TNLYY6 pKa = 9.1PHH8 pKa = 6.85FSAAKK13 pKa = 9.5SLAFQSGIEE22 pKa = 4.0KK23 pKa = 10.43SALVGKK29 pKa = 10.26AVTEE33 pKa = 4.26TVKK36 pKa = 10.92ARR38 pKa = 11.84VNLTRR43 pKa = 11.84LAVEE47 pKa = 4.42TVKK50 pKa = 10.77EE51 pKa = 4.16DD52 pKa = 3.88PLRR55 pKa = 11.84AALTVTSIVSAGWVLKK71 pKa = 10.43KK72 pKa = 10.37AVPFAYY78 pKa = 9.43RR79 pKa = 11.84KK80 pKa = 8.14YY81 pKa = 9.89RR82 pKa = 11.84EE83 pKa = 3.92WRR85 pKa = 11.84EE86 pKa = 3.98RR87 pKa = 11.84PWPVIKK93 pKa = 10.0EE94 pKa = 4.14APSYY98 pKa = 10.11EE99 pKa = 4.24PEE101 pKa = 4.13TAVEE105 pKa = 4.94GSVEE109 pKa = 4.35TNMSFARR116 pKa = 11.84CQGALCVAEE125 pKa = 4.24GQTWKK130 pKa = 9.36TVGGCFRR137 pKa = 11.84LTVGSKK143 pKa = 10.87DD144 pKa = 3.64LLLTAGHH151 pKa = 6.37NLVEE155 pKa = 4.32GEE157 pKa = 4.28RR158 pKa = 11.84YY159 pKa = 9.43AVSRR163 pKa = 11.84MGKK166 pKa = 9.16IKK168 pKa = 10.43EE169 pKa = 4.38LGDD172 pKa = 3.06VWSAKK177 pKa = 10.44DD178 pKa = 3.19ADD180 pKa = 4.24QIYY183 pKa = 10.8SPVTDD188 pKa = 3.91IVAIPLPDD196 pKa = 3.39SFFSGLSMRR205 pKa = 11.84SATTIALPSSTNAQITGALGKK226 pKa = 10.76GSVGKK231 pKa = 8.94LHH233 pKa = 7.22PLGQFGRR240 pKa = 11.84VEE242 pKa = 4.2YY243 pKa = 10.79AGTTAGGYY251 pKa = 9.21SGCPYY256 pKa = 10.26IVGEE260 pKa = 4.24RR261 pKa = 11.84VAAIHH266 pKa = 5.97TNGGRR271 pKa = 11.84KK272 pKa = 9.27NEE274 pKa = 3.82GWEE277 pKa = 3.85IKK279 pKa = 10.5YY280 pKa = 9.76VEE282 pKa = 4.48CLLYY286 pKa = 11.07AHH288 pKa = 6.5FVVSNEE294 pKa = 4.08STVGSEE300 pKa = 3.83DD301 pKa = 3.18AARR304 pKa = 11.84RR305 pKa = 11.84AAQEE309 pKa = 3.87EE310 pKa = 4.59HH311 pKa = 7.48DD312 pKa = 3.99IAPYY316 pKa = 10.8DD317 pKa = 4.26DD318 pKa = 4.89DD319 pKa = 5.97SIIIRR324 pKa = 11.84SRR326 pKa = 11.84KK327 pKa = 6.34TGKK330 pKa = 10.24YY331 pKa = 7.82YY332 pKa = 9.6RR333 pKa = 11.84TRR335 pKa = 11.84EE336 pKa = 3.96TVYY339 pKa = 10.83EE340 pKa = 3.89EE341 pKa = 4.39LMRR344 pKa = 11.84AKK346 pKa = 10.07QSYY349 pKa = 10.38ANNPDD354 pKa = 2.7RR355 pKa = 11.84WADD358 pKa = 3.57QVEE361 pKa = 4.27VDD363 pKa = 5.41LLQQEE368 pKa = 4.68LDD370 pKa = 3.6SYY372 pKa = 10.83MPEE375 pKa = 4.32CASEE379 pKa = 4.28AGNAKK384 pKa = 9.0TPAQAGVGQLDD395 pKa = 3.39GDD397 pKa = 3.86MSVKK401 pKa = 10.3QFLSLLRR408 pKa = 11.84EE409 pKa = 3.87NRR411 pKa = 11.84ALSMSEE417 pKa = 4.07SSEE420 pKa = 3.75QSQLSRR426 pKa = 11.84RR427 pKa = 11.84QRR429 pKa = 11.84RR430 pKa = 11.84QAYY433 pKa = 8.31FGRR436 pKa = 11.84RR437 pKa = 11.84MGLEE441 pKa = 3.48RR442 pKa = 11.84QANIRR447 pKa = 11.84RR448 pKa = 11.84RR449 pKa = 11.84SPTPARR455 pKa = 11.84KK456 pKa = 9.25
Molecular weight: 50.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
809 |
353 |
456 |
404.5 |
45.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.158 ± 2.048 | 1.483 ± 0.145 |
4.821 ± 0.755 | 7.417 ± 0.224 |
3.585 ± 1.014 | 7.664 ± 0.2 |
1.731 ± 0.359 | 3.832 ± 0.09 |
4.944 ± 0.276 | 9.765 ± 1.997 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.472 ± 0.431 | 3.09 ± 0.207 |
4.45 ± 0.245 | 4.203 ± 0.031 |
6.922 ± 0.841 | 6.799 ± 1.328 |
5.315 ± 0.524 | 6.551 ± 0.783 |
2.101 ± 0.49 | 4.697 ± 0.459 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
