
Strigomonas culicis
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Strigomonadinae; Strigomonas
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
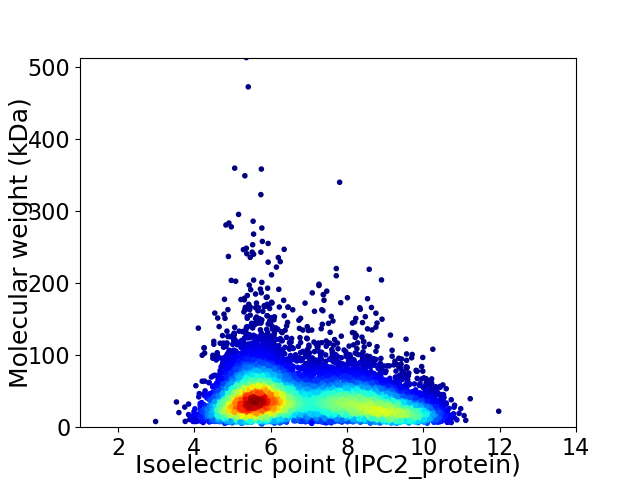
Virtual 2D-PAGE plot for 9986 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
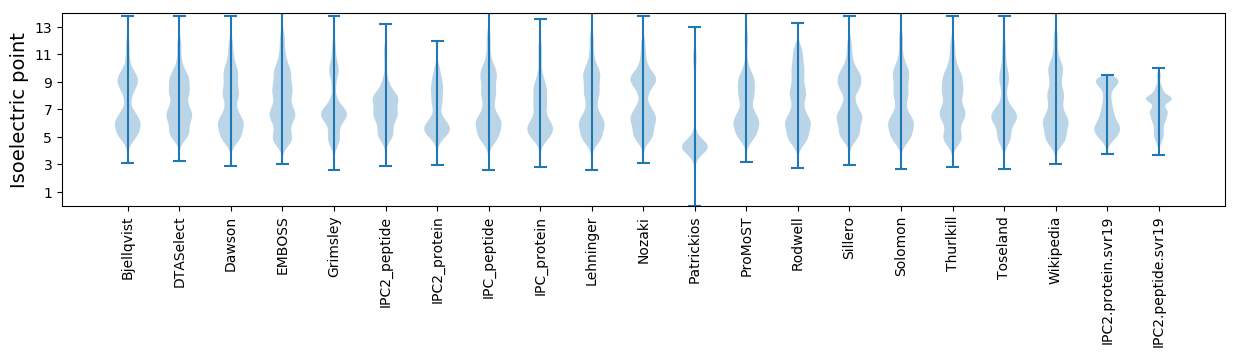
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S9TIR3|S9TIR3_9TRYP Uncharacterized protein OS=Strigomonas culicis OX=28005 GN=STCU_10281 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 5.55APLFNINDD10 pKa = 4.58PLPAPPTNQPPHH22 pKa = 6.07PTSGGSGADD31 pKa = 3.21VDD33 pKa = 5.05PYY35 pKa = 11.33DD36 pKa = 4.22SSGVDD41 pKa = 2.94TADD44 pKa = 4.83RR45 pKa = 11.84SVCCDD50 pKa = 3.37SDD52 pKa = 4.05SSSDD56 pKa = 3.24GSHH59 pKa = 6.92RR60 pKa = 11.84FHH62 pKa = 6.87QLHH65 pKa = 6.32SPQNPTEE72 pKa = 4.25RR73 pKa = 11.84SSMMYY78 pKa = 10.22SSASADD84 pKa = 3.61SLHH87 pKa = 6.53PSSWDD92 pKa = 3.51EE93 pKa = 3.8YY94 pKa = 10.7STDD97 pKa = 3.46SDD99 pKa = 3.9GG100 pKa = 4.63
MM1 pKa = 8.1DD2 pKa = 5.55APLFNINDD10 pKa = 4.58PLPAPPTNQPPHH22 pKa = 6.07PTSGGSGADD31 pKa = 3.21VDD33 pKa = 5.05PYY35 pKa = 11.33DD36 pKa = 4.22SSGVDD41 pKa = 2.94TADD44 pKa = 4.83RR45 pKa = 11.84SVCCDD50 pKa = 3.37SDD52 pKa = 4.05SSSDD56 pKa = 3.24GSHH59 pKa = 6.92RR60 pKa = 11.84FHH62 pKa = 6.87QLHH65 pKa = 6.32SPQNPTEE72 pKa = 4.25RR73 pKa = 11.84SSMMYY78 pKa = 10.22SSASADD84 pKa = 3.61SLHH87 pKa = 6.53PSSWDD92 pKa = 3.51EE93 pKa = 3.8YY94 pKa = 10.7STDD97 pKa = 3.46SDD99 pKa = 3.9GG100 pKa = 4.63
Molecular weight: 10.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S9TEA4|S9TEA4_9TRYP Uncharacterized protein OS=Strigomonas culicis OX=28005 GN=STCU_12149 PE=4 SV=1
MM1 pKa = 7.42SARR4 pKa = 11.84ARR6 pKa = 11.84AAAASVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PTSPRR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84SCRR26 pKa = 11.84ATRR29 pKa = 11.84RR30 pKa = 11.84APRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SPCRR39 pKa = 11.84WPEE42 pKa = 3.58MRR44 pKa = 11.84PRR46 pKa = 11.84QPTYY50 pKa = 10.73CSGSAAAPPRR60 pKa = 11.84WRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PARR67 pKa = 11.84RR68 pKa = 11.84FPGRR72 pKa = 11.84ARR74 pKa = 11.84RR75 pKa = 11.84SPAPWRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84VAVVRR89 pKa = 11.84TASAPRR95 pKa = 11.84SRR97 pKa = 11.84RR98 pKa = 11.84APRR101 pKa = 11.84TAGRR105 pKa = 11.84GCPRR109 pKa = 11.84GRR111 pKa = 11.84ARR113 pKa = 11.84GPSASRR119 pKa = 11.84ARR121 pKa = 11.84RR122 pKa = 11.84AAPPPRR128 pKa = 11.84RR129 pKa = 11.84AAAAGGPPTATRR141 pKa = 11.84GAAAARR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84ATARR153 pKa = 11.84RR154 pKa = 11.84TSARR158 pKa = 11.84SRR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84WSCCCSSGRR171 pKa = 11.84RR172 pKa = 11.84SRR174 pKa = 11.84RR175 pKa = 11.84PRR177 pKa = 11.84PRR179 pKa = 11.84SARR182 pKa = 11.84TRR184 pKa = 11.84RR185 pKa = 11.84PTRR188 pKa = 11.84AARR191 pKa = 11.84TAGGRR196 pKa = 11.84TTRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84APPPSIARR209 pKa = 11.84GTAAIGARR217 pKa = 11.84RR218 pKa = 11.84TTAIGARR225 pKa = 11.84RR226 pKa = 11.84TAAIGARR233 pKa = 11.84RR234 pKa = 11.84TAAISTRR241 pKa = 11.84RR242 pKa = 11.84AAAIRR247 pKa = 11.84ARR249 pKa = 11.84RR250 pKa = 11.84TAAISARR257 pKa = 11.84RR258 pKa = 11.84TAAISARR265 pKa = 11.84RR266 pKa = 11.84TTAPAPAARR275 pKa = 11.84SRR277 pKa = 11.84TAAARR282 pKa = 11.84PPPPVRR288 pKa = 11.84RR289 pKa = 11.84APAAPRR295 pKa = 11.84GSAPRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84RR304 pKa = 11.84TASASGRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84PRR318 pKa = 11.84RR319 pKa = 11.84PPKK322 pKa = 9.84RR323 pKa = 11.84SPRR326 pKa = 11.84RR327 pKa = 11.84AAAPPRR333 pKa = 11.84ARR335 pKa = 11.84APGRR339 pKa = 11.84PPPRR343 pKa = 11.84RR344 pKa = 11.84TSPLRR349 pKa = 11.84CSAASARR356 pKa = 11.84SWTSS360 pKa = 2.74
MM1 pKa = 7.42SARR4 pKa = 11.84ARR6 pKa = 11.84AAAASVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PTSPRR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84SCRR26 pKa = 11.84ATRR29 pKa = 11.84RR30 pKa = 11.84APRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SPCRR39 pKa = 11.84WPEE42 pKa = 3.58MRR44 pKa = 11.84PRR46 pKa = 11.84QPTYY50 pKa = 10.73CSGSAAAPPRR60 pKa = 11.84WRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PARR67 pKa = 11.84RR68 pKa = 11.84FPGRR72 pKa = 11.84ARR74 pKa = 11.84RR75 pKa = 11.84SPAPWRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84VAVVRR89 pKa = 11.84TASAPRR95 pKa = 11.84SRR97 pKa = 11.84RR98 pKa = 11.84APRR101 pKa = 11.84TAGRR105 pKa = 11.84GCPRR109 pKa = 11.84GRR111 pKa = 11.84ARR113 pKa = 11.84GPSASRR119 pKa = 11.84ARR121 pKa = 11.84RR122 pKa = 11.84AAPPPRR128 pKa = 11.84RR129 pKa = 11.84AAAAGGPPTATRR141 pKa = 11.84GAAAARR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84ATARR153 pKa = 11.84RR154 pKa = 11.84TSARR158 pKa = 11.84SRR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84WSCCCSSGRR171 pKa = 11.84RR172 pKa = 11.84SRR174 pKa = 11.84RR175 pKa = 11.84PRR177 pKa = 11.84PRR179 pKa = 11.84SARR182 pKa = 11.84TRR184 pKa = 11.84RR185 pKa = 11.84PTRR188 pKa = 11.84AARR191 pKa = 11.84TAGGRR196 pKa = 11.84TTRR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84APPPSIARR209 pKa = 11.84GTAAIGARR217 pKa = 11.84RR218 pKa = 11.84TTAIGARR225 pKa = 11.84RR226 pKa = 11.84TAAIGARR233 pKa = 11.84RR234 pKa = 11.84TAAISTRR241 pKa = 11.84RR242 pKa = 11.84AAAIRR247 pKa = 11.84ARR249 pKa = 11.84RR250 pKa = 11.84TAAISARR257 pKa = 11.84RR258 pKa = 11.84TAAISARR265 pKa = 11.84RR266 pKa = 11.84TTAPAPAARR275 pKa = 11.84SRR277 pKa = 11.84TAAARR282 pKa = 11.84PPPPVRR288 pKa = 11.84RR289 pKa = 11.84APAAPRR295 pKa = 11.84GSAPRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84RR304 pKa = 11.84TASASGRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84PRR318 pKa = 11.84RR319 pKa = 11.84PPKK322 pKa = 9.84RR323 pKa = 11.84SPRR326 pKa = 11.84RR327 pKa = 11.84AAAPPRR333 pKa = 11.84ARR335 pKa = 11.84APGRR339 pKa = 11.84PPPRR343 pKa = 11.84RR344 pKa = 11.84TSPLRR349 pKa = 11.84CSAASARR356 pKa = 11.84SWTSS360 pKa = 2.74
Molecular weight: 39.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3821174 |
47 |
4543 |
382.7 |
42.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.881 ± 0.041 | 1.868 ± 0.014 |
5.221 ± 0.018 | 6.406 ± 0.025 |
3.878 ± 0.017 | 6.286 ± 0.024 |
2.709 ± 0.013 | 3.972 ± 0.019 |
4.6 ± 0.021 | 9.673 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.43 ± 0.01 | 3.297 ± 0.017 |
5.11 ± 0.024 | 3.954 ± 0.017 |
6.989 ± 0.032 | 6.822 ± 0.03 |
5.589 ± 0.018 | 7.287 ± 0.02 |
1.061 ± 0.008 | 2.915 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
