
bacterium HR24
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
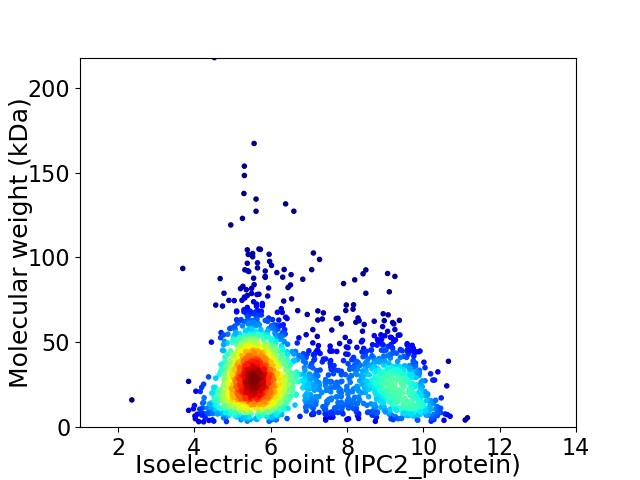
Virtual 2D-PAGE plot for 2129 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
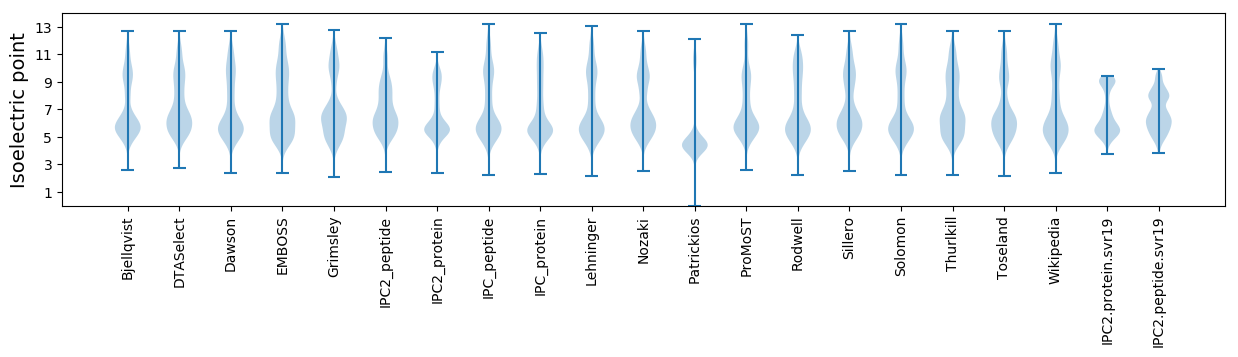
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5YKM3|A0A2H5YKM3_9BACT Benzoylformate decarboxylase OS=bacterium HR24 OX=2035419 GN=mdlC_2 PE=3 SV=1
MM1 pKa = 6.52WRR3 pKa = 11.84SWGRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AGRR12 pKa = 11.84GALFALLVAALVGLPLLAYY31 pKa = 9.59RR32 pKa = 11.84LAGADD37 pKa = 3.77PPTDD41 pKa = 3.36IACDD45 pKa = 3.34IDD47 pKa = 4.28GVGVSYY53 pKa = 10.98GVGFAQDD60 pKa = 3.72PPPPGFRR67 pKa = 11.84VSQVILTGVASSCAGASASVSLYY90 pKa = 8.66STPPPSGTAIASGTATVPTAFSGGSVPVDD119 pKa = 3.31MSPDD123 pKa = 3.55PLAQDD128 pKa = 3.88VNWVEE133 pKa = 4.14VVLAGGTAPTVPEE146 pKa = 4.2CGGIKK151 pKa = 9.81VDD153 pKa = 3.67RR154 pKa = 11.84TYY156 pKa = 11.3SGSDD160 pKa = 3.4GPDD163 pKa = 3.21TITGSNFNDD172 pKa = 3.25LMSGNGGDD180 pKa = 3.71DD181 pKa = 3.3QLYY184 pKa = 10.64GGNNKK189 pKa = 9.82DD190 pKa = 4.21CLLGGPGNDD199 pKa = 2.85RR200 pKa = 11.84LYY202 pKa = 11.33GEE204 pKa = 4.85NGNDD208 pKa = 3.25VLLGGGGNDD217 pKa = 3.33SLYY220 pKa = 11.03GGNGDD225 pKa = 3.7DD226 pKa = 4.19VLYY229 pKa = 10.69GGPGTDD235 pKa = 4.08YY236 pKa = 11.47LDD238 pKa = 4.01GGPGRR243 pKa = 11.84DD244 pKa = 2.74ICYY247 pKa = 8.85VQSGEE252 pKa = 4.33TYY254 pKa = 9.71VNCEE258 pKa = 3.9VVRR261 pKa = 11.84PDD263 pKa = 3.36PTAGGSS269 pKa = 3.23
MM1 pKa = 6.52WRR3 pKa = 11.84SWGRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AGRR12 pKa = 11.84GALFALLVAALVGLPLLAYY31 pKa = 9.59RR32 pKa = 11.84LAGADD37 pKa = 3.77PPTDD41 pKa = 3.36IACDD45 pKa = 3.34IDD47 pKa = 4.28GVGVSYY53 pKa = 10.98GVGFAQDD60 pKa = 3.72PPPPGFRR67 pKa = 11.84VSQVILTGVASSCAGASASVSLYY90 pKa = 8.66STPPPSGTAIASGTATVPTAFSGGSVPVDD119 pKa = 3.31MSPDD123 pKa = 3.55PLAQDD128 pKa = 3.88VNWVEE133 pKa = 4.14VVLAGGTAPTVPEE146 pKa = 4.2CGGIKK151 pKa = 9.81VDD153 pKa = 3.67RR154 pKa = 11.84TYY156 pKa = 11.3SGSDD160 pKa = 3.4GPDD163 pKa = 3.21TITGSNFNDD172 pKa = 3.25LMSGNGGDD180 pKa = 3.71DD181 pKa = 3.3QLYY184 pKa = 10.64GGNNKK189 pKa = 9.82DD190 pKa = 4.21CLLGGPGNDD199 pKa = 2.85RR200 pKa = 11.84LYY202 pKa = 11.33GEE204 pKa = 4.85NGNDD208 pKa = 3.25VLLGGGGNDD217 pKa = 3.33SLYY220 pKa = 11.03GGNGDD225 pKa = 3.7DD226 pKa = 4.19VLYY229 pKa = 10.69GGPGTDD235 pKa = 4.08YY236 pKa = 11.47LDD238 pKa = 4.01GGPGRR243 pKa = 11.84DD244 pKa = 2.74ICYY247 pKa = 8.85VQSGEE252 pKa = 4.33TYY254 pKa = 9.71VNCEE258 pKa = 3.9VVRR261 pKa = 11.84PDD263 pKa = 3.36PTAGGSS269 pKa = 3.23
Molecular weight: 27.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5YH74|A0A2H5YH74_9BACT AsnC_trans_reg domain-containing protein OS=bacterium HR24 OX=2035419 GN=HRbin24_00840 PE=4 SV=1
MM1 pKa = 8.03PKK3 pKa = 8.99RR4 pKa = 11.84TYY6 pKa = 9.29QPKK9 pKa = 9.37NIRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.35RR15 pKa = 11.84KK16 pKa = 8.59HH17 pKa = 5.76GFLARR22 pKa = 11.84MATRR26 pKa = 11.84GGRR29 pKa = 11.84LVLKK33 pKa = 10.38RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.74GRR40 pKa = 11.84WRR42 pKa = 11.84LTVV45 pKa = 3.0
MM1 pKa = 8.03PKK3 pKa = 8.99RR4 pKa = 11.84TYY6 pKa = 9.29QPKK9 pKa = 9.37NIRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.35RR15 pKa = 11.84KK16 pKa = 8.59HH17 pKa = 5.76GFLARR22 pKa = 11.84MATRR26 pKa = 11.84GGRR29 pKa = 11.84LVLKK33 pKa = 10.38RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.74GRR40 pKa = 11.84WRR42 pKa = 11.84LTVV45 pKa = 3.0
Molecular weight: 5.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605099 |
29 |
2083 |
284.2 |
31.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.995 ± 0.075 | 0.998 ± 0.019 |
4.823 ± 0.04 | 6.891 ± 0.065 |
3.049 ± 0.032 | 8.993 ± 0.062 |
2.097 ± 0.028 | 3.68 ± 0.041 |
1.92 ± 0.034 | 11.387 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.143 ± 0.02 | 1.701 ± 0.029 |
6.203 ± 0.044 | 3.442 ± 0.037 |
9.219 ± 0.058 | 4.381 ± 0.033 |
4.199 ± 0.039 | 8.781 ± 0.049 |
1.541 ± 0.027 | 2.556 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
