
Human papillomavirus 151
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 2
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
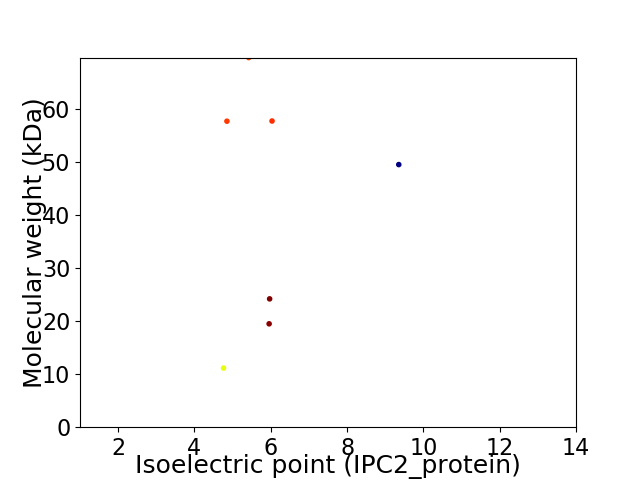
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3VNF8|D3VNF8_9PAPI Replication protein E1 OS=Human papillomavirus 151 OX=743812 GN=E1 PE=3 SV=1
MM1 pKa = 7.54IGKK4 pKa = 8.5QATLCDD10 pKa = 3.55IVLEE14 pKa = 4.14EE15 pKa = 4.64LVLPIDD21 pKa = 3.63LHH23 pKa = 4.95YY24 pKa = 11.06HH25 pKa = 5.86EE26 pKa = 5.87EE27 pKa = 4.19LPEE30 pKa = 4.2LPEE33 pKa = 3.88EE34 pKa = 4.26LEE36 pKa = 4.2EE37 pKa = 4.34VEE39 pKa = 5.05EE40 pKa = 4.29EE41 pKa = 4.78PEE43 pKa = 3.87YY44 pKa = 10.7TPYY47 pKa = 10.79KK48 pKa = 9.94IIVYY52 pKa = 10.32CGGCDD57 pKa = 3.17TKK59 pKa = 11.37LKK61 pKa = 10.35LHH63 pKa = 6.11VLATLSGIRR72 pKa = 11.84DD73 pKa = 4.02FQASLLGPVKK83 pKa = 10.18LLCPSCRR90 pKa = 11.84EE91 pKa = 3.84EE92 pKa = 3.77IRR94 pKa = 11.84NGRR97 pKa = 11.84RR98 pKa = 3.02
MM1 pKa = 7.54IGKK4 pKa = 8.5QATLCDD10 pKa = 3.55IVLEE14 pKa = 4.14EE15 pKa = 4.64LVLPIDD21 pKa = 3.63LHH23 pKa = 4.95YY24 pKa = 11.06HH25 pKa = 5.86EE26 pKa = 5.87EE27 pKa = 4.19LPEE30 pKa = 4.2LPEE33 pKa = 3.88EE34 pKa = 4.26LEE36 pKa = 4.2EE37 pKa = 4.34VEE39 pKa = 5.05EE40 pKa = 4.29EE41 pKa = 4.78PEE43 pKa = 3.87YY44 pKa = 10.7TPYY47 pKa = 10.79KK48 pKa = 9.94IIVYY52 pKa = 10.32CGGCDD57 pKa = 3.17TKK59 pKa = 11.37LKK61 pKa = 10.35LHH63 pKa = 6.11VLATLSGIRR72 pKa = 11.84DD73 pKa = 4.02FQASLLGPVKK83 pKa = 10.18LLCPSCRR90 pKa = 11.84EE91 pKa = 3.84EE92 pKa = 3.77IRR94 pKa = 11.84NGRR97 pKa = 11.84RR98 pKa = 3.02
Molecular weight: 11.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3VNG0|D3VNG0_9PAPI Putative E4 product OS=Human papillomavirus 151 OX=743812 GN=E4 PE=4 SV=1
MM1 pKa = 7.64EE2 pKa = 5.49KK3 pKa = 10.66LNEE6 pKa = 4.15RR7 pKa = 11.84FSALQEE13 pKa = 4.13KK14 pKa = 10.62LMDD17 pKa = 4.3LYY19 pKa = 11.28EE20 pKa = 5.14SGVEE24 pKa = 3.96DD25 pKa = 5.76LEE27 pKa = 4.52TQIQHH32 pKa = 5.68WKK34 pKa = 10.34LLRR37 pKa = 11.84QEE39 pKa = 4.47QIIFYY44 pKa = 7.62YY45 pKa = 10.39ARR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 5.25GIMRR53 pKa = 11.84LGYY56 pKa = 10.17QPVPPLATSEE66 pKa = 4.58SKK68 pKa = 11.13AKK70 pKa = 10.43DD71 pKa = 3.81AIAMGLLLEE80 pKa = 4.68SLQKK84 pKa = 10.89SKK86 pKa = 10.77FSEE89 pKa = 4.55EE90 pKa = 3.54PWTLVEE96 pKa = 4.34TSLEE100 pKa = 4.45TVRR103 pKa = 11.84SPPADD108 pKa = 3.59CFEE111 pKa = 4.94KK112 pKa = 10.69GPKK115 pKa = 9.47SVEE118 pKa = 4.01VYY120 pKa = 10.71FDD122 pKa = 3.98GDD124 pKa = 3.74PEE126 pKa = 4.3NVMSYY131 pKa = 7.39TVWTYY136 pKa = 10.83IYY138 pKa = 10.2YY139 pKa = 8.53QTDD142 pKa = 3.34DD143 pKa = 3.75EE144 pKa = 4.61TWAKK148 pKa = 10.79VEE150 pKa = 3.9GHH152 pKa = 5.76VDD154 pKa = 3.42YY155 pKa = 11.48AGAYY159 pKa = 8.77YY160 pKa = 9.79MEE162 pKa = 4.89GTFKK166 pKa = 10.29TYY168 pKa = 10.4YY169 pKa = 10.05IKK171 pKa = 10.91FEE173 pKa = 4.25TDD175 pKa = 2.4AKK177 pKa = 10.65RR178 pKa = 11.84YY179 pKa = 6.65GTTGYY184 pKa = 10.19WEE186 pKa = 3.87VHH188 pKa = 4.75VNKK191 pKa = 9.35DD192 pKa = 3.53TVFTPVTSSTPPAGVASQNPASEE215 pKa = 4.51PVSTSDD221 pKa = 3.29PAKK224 pKa = 9.04RR225 pKa = 11.84TSQATHH231 pKa = 4.52QRR233 pKa = 11.84YY234 pKa = 8.65GRR236 pKa = 11.84KK237 pKa = 9.43ASSPTVTSIRR247 pKa = 11.84AQKK250 pKa = 9.75RR251 pKa = 11.84RR252 pKa = 11.84QRR254 pKa = 11.84QRR256 pKa = 11.84QEE258 pKa = 3.52TPTRR262 pKa = 11.84QRR264 pKa = 11.84TNRR267 pKa = 11.84SRR269 pKa = 11.84SRR271 pKa = 11.84STEE274 pKa = 3.54RR275 pKa = 11.84RR276 pKa = 11.84GGRR279 pKa = 11.84ATRR282 pKa = 11.84RR283 pKa = 11.84SLSRR287 pKa = 11.84GSSKK291 pKa = 10.54SPRR294 pKa = 11.84RR295 pKa = 11.84GGRR298 pKa = 11.84SGGGPLTRR306 pKa = 11.84SRR308 pKa = 11.84SRR310 pKa = 11.84SRR312 pKa = 11.84SLTRR316 pKa = 11.84KK317 pKa = 8.35SANGGGVAPDD327 pKa = 3.36EE328 pKa = 4.47VGATVRR334 pKa = 11.84SISRR338 pKa = 11.84QHH340 pKa = 6.45SGRR343 pKa = 11.84LAQLLDD349 pKa = 3.62AAKK352 pKa = 9.99DD353 pKa = 3.57PPVILLRR360 pKa = 11.84GASNTLKK367 pKa = 10.38CYY369 pKa = 10.33RR370 pKa = 11.84YY371 pKa = 10.07RR372 pKa = 11.84FRR374 pKa = 11.84KK375 pKa = 8.72KK376 pKa = 9.81HH377 pKa = 5.83AGNFQLISTTWSWVGGHH394 pKa = 4.66TTDD397 pKa = 3.84RR398 pKa = 11.84IGRR401 pKa = 11.84ARR403 pKa = 11.84MLISFHH409 pKa = 6.87SNSEE413 pKa = 4.17RR414 pKa = 11.84EE415 pKa = 3.99TCLQQMKK422 pKa = 10.51LPLGVEE428 pKa = 4.08WSYY431 pKa = 11.91GQFDD435 pKa = 4.19DD436 pKa = 5.18LL437 pKa = 5.54
MM1 pKa = 7.64EE2 pKa = 5.49KK3 pKa = 10.66LNEE6 pKa = 4.15RR7 pKa = 11.84FSALQEE13 pKa = 4.13KK14 pKa = 10.62LMDD17 pKa = 4.3LYY19 pKa = 11.28EE20 pKa = 5.14SGVEE24 pKa = 3.96DD25 pKa = 5.76LEE27 pKa = 4.52TQIQHH32 pKa = 5.68WKK34 pKa = 10.34LLRR37 pKa = 11.84QEE39 pKa = 4.47QIIFYY44 pKa = 7.62YY45 pKa = 10.39ARR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 5.25GIMRR53 pKa = 11.84LGYY56 pKa = 10.17QPVPPLATSEE66 pKa = 4.58SKK68 pKa = 11.13AKK70 pKa = 10.43DD71 pKa = 3.81AIAMGLLLEE80 pKa = 4.68SLQKK84 pKa = 10.89SKK86 pKa = 10.77FSEE89 pKa = 4.55EE90 pKa = 3.54PWTLVEE96 pKa = 4.34TSLEE100 pKa = 4.45TVRR103 pKa = 11.84SPPADD108 pKa = 3.59CFEE111 pKa = 4.94KK112 pKa = 10.69GPKK115 pKa = 9.47SVEE118 pKa = 4.01VYY120 pKa = 10.71FDD122 pKa = 3.98GDD124 pKa = 3.74PEE126 pKa = 4.3NVMSYY131 pKa = 7.39TVWTYY136 pKa = 10.83IYY138 pKa = 10.2YY139 pKa = 8.53QTDD142 pKa = 3.34DD143 pKa = 3.75EE144 pKa = 4.61TWAKK148 pKa = 10.79VEE150 pKa = 3.9GHH152 pKa = 5.76VDD154 pKa = 3.42YY155 pKa = 11.48AGAYY159 pKa = 8.77YY160 pKa = 9.79MEE162 pKa = 4.89GTFKK166 pKa = 10.29TYY168 pKa = 10.4YY169 pKa = 10.05IKK171 pKa = 10.91FEE173 pKa = 4.25TDD175 pKa = 2.4AKK177 pKa = 10.65RR178 pKa = 11.84YY179 pKa = 6.65GTTGYY184 pKa = 10.19WEE186 pKa = 3.87VHH188 pKa = 4.75VNKK191 pKa = 9.35DD192 pKa = 3.53TVFTPVTSSTPPAGVASQNPASEE215 pKa = 4.51PVSTSDD221 pKa = 3.29PAKK224 pKa = 9.04RR225 pKa = 11.84TSQATHH231 pKa = 4.52QRR233 pKa = 11.84YY234 pKa = 8.65GRR236 pKa = 11.84KK237 pKa = 9.43ASSPTVTSIRR247 pKa = 11.84AQKK250 pKa = 9.75RR251 pKa = 11.84RR252 pKa = 11.84QRR254 pKa = 11.84QRR256 pKa = 11.84QEE258 pKa = 3.52TPTRR262 pKa = 11.84QRR264 pKa = 11.84TNRR267 pKa = 11.84SRR269 pKa = 11.84SRR271 pKa = 11.84STEE274 pKa = 3.54RR275 pKa = 11.84RR276 pKa = 11.84GGRR279 pKa = 11.84ATRR282 pKa = 11.84RR283 pKa = 11.84SLSRR287 pKa = 11.84GSSKK291 pKa = 10.54SPRR294 pKa = 11.84RR295 pKa = 11.84GGRR298 pKa = 11.84SGGGPLTRR306 pKa = 11.84SRR308 pKa = 11.84SRR310 pKa = 11.84SRR312 pKa = 11.84SLTRR316 pKa = 11.84KK317 pKa = 8.35SANGGGVAPDD327 pKa = 3.36EE328 pKa = 4.47VGATVRR334 pKa = 11.84SISRR338 pKa = 11.84QHH340 pKa = 6.45SGRR343 pKa = 11.84LAQLLDD349 pKa = 3.62AAKK352 pKa = 9.99DD353 pKa = 3.57PPVILLRR360 pKa = 11.84GASNTLKK367 pKa = 10.38CYY369 pKa = 10.33RR370 pKa = 11.84YY371 pKa = 10.07RR372 pKa = 11.84FRR374 pKa = 11.84KK375 pKa = 8.72KK376 pKa = 9.81HH377 pKa = 5.83AGNFQLISTTWSWVGGHH394 pKa = 4.66TTDD397 pKa = 3.84RR398 pKa = 11.84IGRR401 pKa = 11.84ARR403 pKa = 11.84MLISFHH409 pKa = 6.87SNSEE413 pKa = 4.17RR414 pKa = 11.84EE415 pKa = 3.99TCLQQMKK422 pKa = 10.51LPLGVEE428 pKa = 4.08WSYY431 pKa = 11.91GQFDD435 pKa = 4.19DD436 pKa = 5.18LL437 pKa = 5.54
Molecular weight: 49.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2557 |
98 |
608 |
365.3 |
41.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.553 ± 0.524 | 2.034 ± 0.564 |
6.218 ± 0.467 | 6.727 ± 0.639 |
4.106 ± 0.599 | 6.101 ± 0.527 |
2.307 ± 0.21 | 5.084 ± 0.615 |
4.967 ± 0.753 | 8.76 ± 0.779 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.034 ± 0.429 | 3.442 ± 0.671 |
6.453 ± 1.146 | 5.084 ± 0.693 |
6.766 ± 0.81 | 7.587 ± 0.737 |
6.101 ± 0.988 | 5.866 ± 0.679 |
1.369 ± 0.311 | 3.442 ± 0.424 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
