
Bivalve hepelivirus G
Taxonomy: Viruses; Riboviria; unclassified Riboviria
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
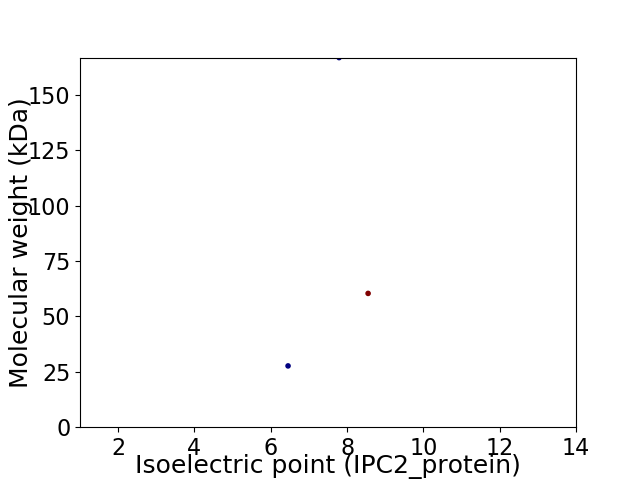
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L4FQZ1|A0A1L4FQZ1_9VIRU Replicative protein OS=Bivalve hepelivirus G OX=1926998 PE=4 SV=1
MM1 pKa = 7.78DD2 pKa = 3.7VSHH5 pKa = 6.65TKK7 pKa = 10.88SMLEE11 pKa = 4.13LEE13 pKa = 4.67LEE15 pKa = 4.36LFGMLGVNQKK25 pKa = 10.5IIDD28 pKa = 4.71FYY30 pKa = 11.37ATMRR34 pKa = 11.84TKK36 pKa = 9.37WCNLYY41 pKa = 9.97QCRR44 pKa = 11.84EE45 pKa = 4.47GIAMLHH51 pKa = 5.59AWYY54 pKa = 8.2MQHH57 pKa = 6.73SGQPLTLTGNTLLNMAVIGFAYY79 pKa = 10.12RR80 pKa = 11.84IVGMLYY86 pKa = 10.56GAFKK90 pKa = 11.04GDD92 pKa = 3.94DD93 pKa = 2.99SHH95 pKa = 6.84MRR97 pKa = 11.84AKK99 pKa = 10.46KK100 pKa = 9.83ISTVPGRR107 pKa = 11.84TTQAYY112 pKa = 10.07ADD114 pKa = 3.71YY115 pKa = 10.1GYY117 pKa = 10.99KK118 pKa = 10.53LKK120 pKa = 10.65ISFEE124 pKa = 4.23KK125 pKa = 10.72VSEE128 pKa = 4.92FIANFVTPYY137 pKa = 11.04GFFPDD142 pKa = 3.49VVRR145 pKa = 11.84RR146 pKa = 11.84SVKK149 pKa = 10.09AVSKK153 pKa = 10.19IYY155 pKa = 10.19EE156 pKa = 4.44DD157 pKa = 3.69EE158 pKa = 4.4EE159 pKa = 4.16SWEE162 pKa = 4.02EE163 pKa = 3.78SRR165 pKa = 11.84VNLKK169 pKa = 10.16EE170 pKa = 3.76VLSVVNSADD179 pKa = 3.32KK180 pKa = 11.16YY181 pKa = 11.19KK182 pKa = 10.66IGIDD186 pKa = 3.56CAYY189 pKa = 10.7LHH191 pKa = 6.27YY192 pKa = 9.77RR193 pKa = 11.84DD194 pKa = 4.39KK195 pKa = 11.47GVAITRR201 pKa = 11.84EE202 pKa = 4.23QVSVLYY208 pKa = 10.31QYY210 pKa = 10.9LIQLSNTKK218 pKa = 10.3YY219 pKa = 10.92ADD221 pKa = 3.31AEE223 pKa = 4.61FIQSEE228 pKa = 4.2DD229 pKa = 3.22RR230 pKa = 11.84LTYY233 pKa = 10.34SDD235 pKa = 4.66NYY237 pKa = 9.86QSRR240 pKa = 3.68
MM1 pKa = 7.78DD2 pKa = 3.7VSHH5 pKa = 6.65TKK7 pKa = 10.88SMLEE11 pKa = 4.13LEE13 pKa = 4.67LEE15 pKa = 4.36LFGMLGVNQKK25 pKa = 10.5IIDD28 pKa = 4.71FYY30 pKa = 11.37ATMRR34 pKa = 11.84TKK36 pKa = 9.37WCNLYY41 pKa = 9.97QCRR44 pKa = 11.84EE45 pKa = 4.47GIAMLHH51 pKa = 5.59AWYY54 pKa = 8.2MQHH57 pKa = 6.73SGQPLTLTGNTLLNMAVIGFAYY79 pKa = 10.12RR80 pKa = 11.84IVGMLYY86 pKa = 10.56GAFKK90 pKa = 11.04GDD92 pKa = 3.94DD93 pKa = 2.99SHH95 pKa = 6.84MRR97 pKa = 11.84AKK99 pKa = 10.46KK100 pKa = 9.83ISTVPGRR107 pKa = 11.84TTQAYY112 pKa = 10.07ADD114 pKa = 3.71YY115 pKa = 10.1GYY117 pKa = 10.99KK118 pKa = 10.53LKK120 pKa = 10.65ISFEE124 pKa = 4.23KK125 pKa = 10.72VSEE128 pKa = 4.92FIANFVTPYY137 pKa = 11.04GFFPDD142 pKa = 3.49VVRR145 pKa = 11.84RR146 pKa = 11.84SVKK149 pKa = 10.09AVSKK153 pKa = 10.19IYY155 pKa = 10.19EE156 pKa = 4.44DD157 pKa = 3.69EE158 pKa = 4.4EE159 pKa = 4.16SWEE162 pKa = 4.02EE163 pKa = 3.78SRR165 pKa = 11.84VNLKK169 pKa = 10.16EE170 pKa = 3.76VLSVVNSADD179 pKa = 3.32KK180 pKa = 11.16YY181 pKa = 11.19KK182 pKa = 10.66IGIDD186 pKa = 3.56CAYY189 pKa = 10.7LHH191 pKa = 6.27YY192 pKa = 9.77RR193 pKa = 11.84DD194 pKa = 4.39KK195 pKa = 11.47GVAITRR201 pKa = 11.84EE202 pKa = 4.23QVSVLYY208 pKa = 10.31QYY210 pKa = 10.9LIQLSNTKK218 pKa = 10.3YY219 pKa = 10.92ADD221 pKa = 3.31AEE223 pKa = 4.61FIQSEE228 pKa = 4.2DD229 pKa = 3.22RR230 pKa = 11.84LTYY233 pKa = 10.34SDD235 pKa = 4.66NYY237 pKa = 9.86QSRR240 pKa = 3.68
Molecular weight: 27.67 kDa
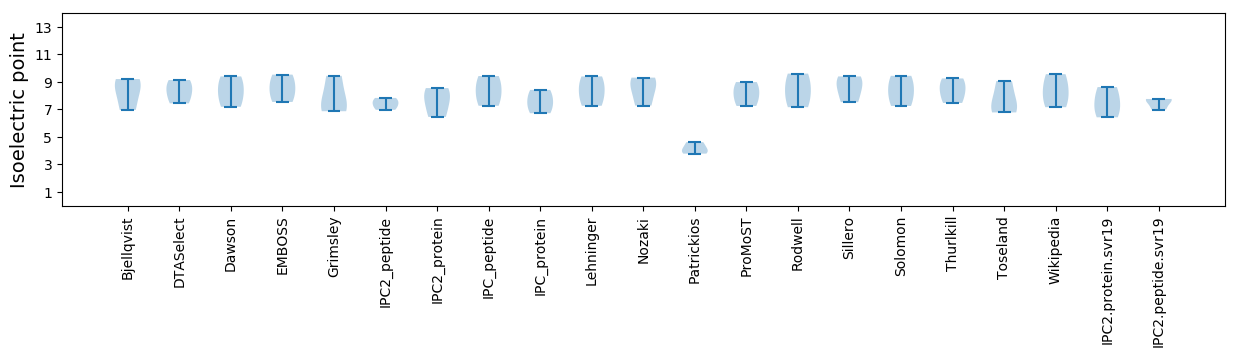
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L4FQZ2|A0A1L4FQZ2_9VIRU Structural protein OS=Bivalve hepelivirus G OX=1926998 PE=4 SV=1
MM1 pKa = 7.11NAAAPLVDD9 pKa = 3.34NSMNFDD15 pKa = 3.75PMSDD19 pKa = 3.39STSMPEE25 pKa = 3.78QTHH28 pKa = 6.23GKK30 pKa = 7.53TLTPSQAFVCKK41 pKa = 8.95VTHH44 pKa = 6.89PPTTVPEE51 pKa = 4.26FAGLPTQDD59 pKa = 2.57TRR61 pKa = 11.84TQVVYY66 pKa = 11.08NMRR69 pKa = 11.84NIDD72 pKa = 3.99VIKK75 pKa = 10.11TPITFDD81 pKa = 3.23KK82 pKa = 10.99TSGKK86 pKa = 10.24YY87 pKa = 7.82EE88 pKa = 3.94QNSWTDD94 pKa = 3.08HH95 pKa = 5.97EE96 pKa = 5.25DD97 pKa = 3.25YY98 pKa = 11.47SFIVPTGARR107 pKa = 11.84IKK109 pKa = 10.35WFGCCYY115 pKa = 9.78QVNDD119 pKa = 3.98SQTTPTTADD128 pKa = 4.11AYY130 pKa = 10.8DD131 pKa = 3.4QDD133 pKa = 4.14LANVGVQDD141 pKa = 4.12NFDD144 pKa = 3.87FKK146 pKa = 11.16SWSSTVNLYY155 pKa = 10.19RR156 pKa = 11.84PCYY159 pKa = 10.14KK160 pKa = 10.65SITLYY165 pKa = 10.75PNVTAFNNQGIISAQQFNPNILFSGSTSALSYY197 pKa = 10.66DD198 pKa = 3.52KK199 pKa = 10.94PKK201 pKa = 11.27LFMQALDD208 pKa = 3.86HH209 pKa = 6.95LYY211 pKa = 10.86SQINDD216 pKa = 3.34KK217 pKa = 10.82LFCNEE222 pKa = 3.56EE223 pKa = 4.1THH225 pKa = 7.27VDD227 pKa = 3.77FHH229 pKa = 5.31QTVVEE234 pKa = 4.35SWFKK238 pKa = 9.52TRR240 pKa = 11.84KK241 pKa = 9.29LRR243 pKa = 11.84VRR245 pKa = 11.84GLNLDD250 pKa = 3.89PNNFLQILNIGNVGYY265 pKa = 10.89GSDD268 pKa = 3.55TSSLVPTPSQIAQNSMRR285 pKa = 11.84SYY287 pKa = 10.5QDD289 pKa = 3.01KK290 pKa = 10.36FINGAFVVSRR300 pKa = 11.84INTLSPKK307 pKa = 9.28WMAGSNTNIPVNGLYY322 pKa = 10.15EE323 pKa = 4.07CWLYY327 pKa = 11.28SIGPDD332 pKa = 3.49GRR334 pKa = 11.84PHH336 pKa = 7.08LVSLKK341 pKa = 10.78DD342 pKa = 3.37PVAAGTAAADD352 pKa = 4.02AKK354 pKa = 11.05PMLDD358 pKa = 3.91TLWSSDD364 pKa = 3.37MTWQFIRR371 pKa = 11.84VQGISPNVTASTAPAVVSPIAIKK394 pKa = 10.11HH395 pKa = 4.93YY396 pKa = 10.55YY397 pKa = 9.52GIEE400 pKa = 3.99AQPVWNGPWNGIARR414 pKa = 11.84MSPKK418 pKa = 9.98PSLSEE423 pKa = 3.72MQALMDD429 pKa = 4.31TFYY432 pKa = 11.07EE433 pKa = 4.51MPDD436 pKa = 3.41AMPAKK441 pKa = 8.78YY442 pKa = 10.37NAMGAFLPFLASALPHH458 pKa = 6.5ALRR461 pKa = 11.84FVKK464 pKa = 10.51DD465 pKa = 4.46LITSKK470 pKa = 10.54KK471 pKa = 9.63SKK473 pKa = 10.61NDD475 pKa = 3.13GTVGVATTTKK485 pKa = 10.12RR486 pKa = 11.84SKK488 pKa = 11.13ASTKK492 pKa = 10.2PISSNNDD499 pKa = 2.68KK500 pKa = 11.08QIIANLRR507 pKa = 11.84KK508 pKa = 9.9QIASMQLNNRR518 pKa = 11.84PPSKK522 pKa = 9.92RR523 pKa = 11.84RR524 pKa = 11.84KK525 pKa = 6.46TRR527 pKa = 11.84KK528 pKa = 8.83PRR530 pKa = 11.84SKK532 pKa = 10.39SSPIIINEE540 pKa = 3.88RR541 pKa = 11.84TKK543 pKa = 11.25
MM1 pKa = 7.11NAAAPLVDD9 pKa = 3.34NSMNFDD15 pKa = 3.75PMSDD19 pKa = 3.39STSMPEE25 pKa = 3.78QTHH28 pKa = 6.23GKK30 pKa = 7.53TLTPSQAFVCKK41 pKa = 8.95VTHH44 pKa = 6.89PPTTVPEE51 pKa = 4.26FAGLPTQDD59 pKa = 2.57TRR61 pKa = 11.84TQVVYY66 pKa = 11.08NMRR69 pKa = 11.84NIDD72 pKa = 3.99VIKK75 pKa = 10.11TPITFDD81 pKa = 3.23KK82 pKa = 10.99TSGKK86 pKa = 10.24YY87 pKa = 7.82EE88 pKa = 3.94QNSWTDD94 pKa = 3.08HH95 pKa = 5.97EE96 pKa = 5.25DD97 pKa = 3.25YY98 pKa = 11.47SFIVPTGARR107 pKa = 11.84IKK109 pKa = 10.35WFGCCYY115 pKa = 9.78QVNDD119 pKa = 3.98SQTTPTTADD128 pKa = 4.11AYY130 pKa = 10.8DD131 pKa = 3.4QDD133 pKa = 4.14LANVGVQDD141 pKa = 4.12NFDD144 pKa = 3.87FKK146 pKa = 11.16SWSSTVNLYY155 pKa = 10.19RR156 pKa = 11.84PCYY159 pKa = 10.14KK160 pKa = 10.65SITLYY165 pKa = 10.75PNVTAFNNQGIISAQQFNPNILFSGSTSALSYY197 pKa = 10.66DD198 pKa = 3.52KK199 pKa = 10.94PKK201 pKa = 11.27LFMQALDD208 pKa = 3.86HH209 pKa = 6.95LYY211 pKa = 10.86SQINDD216 pKa = 3.34KK217 pKa = 10.82LFCNEE222 pKa = 3.56EE223 pKa = 4.1THH225 pKa = 7.27VDD227 pKa = 3.77FHH229 pKa = 5.31QTVVEE234 pKa = 4.35SWFKK238 pKa = 9.52TRR240 pKa = 11.84KK241 pKa = 9.29LRR243 pKa = 11.84VRR245 pKa = 11.84GLNLDD250 pKa = 3.89PNNFLQILNIGNVGYY265 pKa = 10.89GSDD268 pKa = 3.55TSSLVPTPSQIAQNSMRR285 pKa = 11.84SYY287 pKa = 10.5QDD289 pKa = 3.01KK290 pKa = 10.36FINGAFVVSRR300 pKa = 11.84INTLSPKK307 pKa = 9.28WMAGSNTNIPVNGLYY322 pKa = 10.15EE323 pKa = 4.07CWLYY327 pKa = 11.28SIGPDD332 pKa = 3.49GRR334 pKa = 11.84PHH336 pKa = 7.08LVSLKK341 pKa = 10.78DD342 pKa = 3.37PVAAGTAAADD352 pKa = 4.02AKK354 pKa = 11.05PMLDD358 pKa = 3.91TLWSSDD364 pKa = 3.37MTWQFIRR371 pKa = 11.84VQGISPNVTASTAPAVVSPIAIKK394 pKa = 10.11HH395 pKa = 4.93YY396 pKa = 10.55YY397 pKa = 9.52GIEE400 pKa = 3.99AQPVWNGPWNGIARR414 pKa = 11.84MSPKK418 pKa = 9.98PSLSEE423 pKa = 3.72MQALMDD429 pKa = 4.31TFYY432 pKa = 11.07EE433 pKa = 4.51MPDD436 pKa = 3.41AMPAKK441 pKa = 8.78YY442 pKa = 10.37NAMGAFLPFLASALPHH458 pKa = 6.5ALRR461 pKa = 11.84FVKK464 pKa = 10.51DD465 pKa = 4.46LITSKK470 pKa = 10.54KK471 pKa = 9.63SKK473 pKa = 10.61NDD475 pKa = 3.13GTVGVATTTKK485 pKa = 10.12RR486 pKa = 11.84SKK488 pKa = 11.13ASTKK492 pKa = 10.2PISSNNDD499 pKa = 2.68KK500 pKa = 11.08QIIANLRR507 pKa = 11.84KK508 pKa = 9.9QIASMQLNNRR518 pKa = 11.84PPSKK522 pKa = 9.92RR523 pKa = 11.84RR524 pKa = 11.84KK525 pKa = 6.46TRR527 pKa = 11.84KK528 pKa = 8.83PRR530 pKa = 11.84SKK532 pKa = 10.39SSPIIINEE540 pKa = 3.88RR541 pKa = 11.84TKK543 pKa = 11.25
Molecular weight: 60.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2236 |
240 |
1453 |
745.3 |
84.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.903 ± 0.677 | 1.699 ± 0.295 |
6.127 ± 0.276 | 4.338 ± 0.824 |
4.249 ± 0.022 | 4.383 ± 0.383 |
2.326 ± 0.295 | 7.469 ± 0.921 |
7.737 ± 0.725 | 7.2 ± 0.382 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.728 ± 0.335 | 7.111 ± 0.784 |
3.98 ± 1.29 | 3.488 ± 0.601 |
4.07 ± 0.206 | 7.335 ± 0.848 |
6.664 ± 0.555 | 6.798 ± 0.346 |
0.984 ± 0.374 | 5.411 ± 0.822 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
