
Salibacter halophilus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Salibacteraceae; Salibacter
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
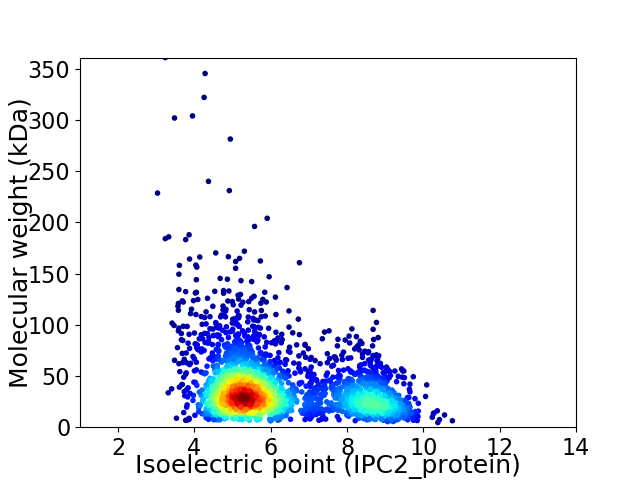
Virtual 2D-PAGE plot for 2675 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N6M6E2|A0A6N6M6E2_9FLAO Short-chain fatty acid transporter OS=Salibacter halophilus OX=1803916 GN=F3059_09645 PE=4 SV=1
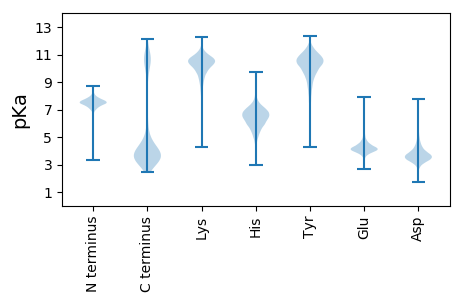
II1 pKa = 7.43TDD3 pKa = 3.94GSNNVSTSTAIVTVEE18 pKa = 3.89DD19 pKa = 4.52TIAPNIQTQSVTLYY33 pKa = 10.78LDD35 pKa = 3.55SLGQASTTAMSIDD48 pKa = 3.77AGSSDD53 pKa = 3.54NCAIDD58 pKa = 3.88TMFLSEE64 pKa = 4.46NNFDD68 pKa = 4.14CSHH71 pKa = 6.98IGTNTVQLIATDD83 pKa = 3.79VNGNTDD89 pKa = 2.78SSSAVVTVEE98 pKa = 3.7DD99 pKa = 4.56TIAPEE104 pKa = 4.44VITQGVTLYY113 pKa = 10.99LDD115 pKa = 3.61STGTAVLTPSVFDD128 pKa = 3.92NGSNDD133 pKa = 3.17NCSIDD138 pKa = 4.63SMSLSQDD145 pKa = 2.84TFTCSDD151 pKa = 3.63LGLHH155 pKa = 6.0VVDD158 pKa = 5.57LHH160 pKa = 6.33VTDD163 pKa = 3.97QSGNGATSNALVSVTDD179 pKa = 4.42TIAPLLRR186 pKa = 11.84SFSNITVYY194 pKa = 10.96LDD196 pKa = 3.36SSGTASIDD204 pKa = 3.92AGMLDD209 pKa = 4.03SNSVDD214 pKa = 3.14NCSLDD219 pKa = 3.55TIYY222 pKa = 11.09LSRR225 pKa = 11.84TTLGCNDD232 pKa = 4.1LGSTTVEE239 pKa = 4.56LIGKK243 pKa = 8.6DD244 pKa = 3.06GSMNEE249 pKa = 3.72SRR251 pKa = 11.84ILVNLTVEE259 pKa = 4.39DD260 pKa = 3.98TLAPTIEE267 pKa = 4.74CPSSFDD273 pKa = 3.13ACAGIVNFPDD283 pKa = 3.66AVSNDD288 pKa = 3.26NCTSTVTNTGNVFSGDD304 pKa = 3.44SLTEE308 pKa = 3.97GEE310 pKa = 4.59YY311 pKa = 9.21VTSFEE316 pKa = 4.83AVDD319 pKa = 3.91GSGNVASCTTSFSVNAIPEE338 pKa = 4.04VDD340 pKa = 4.24LGADD344 pKa = 3.43TAVGPGHH351 pKa = 5.93VVSFSVNDD359 pKa = 3.88TVSEE363 pKa = 4.32YY364 pKa = 10.96LWSTGDD370 pKa = 3.37TTRR373 pKa = 11.84SLDD376 pKa = 3.92LKK378 pKa = 10.87VLTDD382 pKa = 3.31TTVWISVTDD391 pKa = 3.93SNSCTGSDD399 pKa = 3.31TATVTTLTSIKK410 pKa = 10.03RR411 pKa = 11.84SKK413 pKa = 9.57KK414 pKa = 7.21TVNVSVYY421 pKa = 9.71PNPTRR426 pKa = 11.84GEE428 pKa = 4.19VKK430 pKa = 10.5VEE432 pKa = 3.7LLGVEE437 pKa = 4.12NLNYY441 pKa = 10.43HH442 pKa = 7.2LIDD445 pKa = 3.31MAGRR449 pKa = 11.84ILQSGNMQNGLNRR462 pKa = 11.84IDD464 pKa = 3.85LSRR467 pKa = 11.84YY468 pKa = 6.07ATSVYY473 pKa = 10.47YY474 pKa = 10.63LQLQNEE480 pKa = 4.3QGEE483 pKa = 4.96GIKK486 pKa = 10.7VMRR489 pKa = 11.84LVKK492 pKa = 10.34QQ493 pKa = 3.31
II1 pKa = 7.43TDD3 pKa = 3.94GSNNVSTSTAIVTVEE18 pKa = 3.89DD19 pKa = 4.52TIAPNIQTQSVTLYY33 pKa = 10.78LDD35 pKa = 3.55SLGQASTTAMSIDD48 pKa = 3.77AGSSDD53 pKa = 3.54NCAIDD58 pKa = 3.88TMFLSEE64 pKa = 4.46NNFDD68 pKa = 4.14CSHH71 pKa = 6.98IGTNTVQLIATDD83 pKa = 3.79VNGNTDD89 pKa = 2.78SSSAVVTVEE98 pKa = 3.7DD99 pKa = 4.56TIAPEE104 pKa = 4.44VITQGVTLYY113 pKa = 10.99LDD115 pKa = 3.61STGTAVLTPSVFDD128 pKa = 3.92NGSNDD133 pKa = 3.17NCSIDD138 pKa = 4.63SMSLSQDD145 pKa = 2.84TFTCSDD151 pKa = 3.63LGLHH155 pKa = 6.0VVDD158 pKa = 5.57LHH160 pKa = 6.33VTDD163 pKa = 3.97QSGNGATSNALVSVTDD179 pKa = 4.42TIAPLLRR186 pKa = 11.84SFSNITVYY194 pKa = 10.96LDD196 pKa = 3.36SSGTASIDD204 pKa = 3.92AGMLDD209 pKa = 4.03SNSVDD214 pKa = 3.14NCSLDD219 pKa = 3.55TIYY222 pKa = 11.09LSRR225 pKa = 11.84TTLGCNDD232 pKa = 4.1LGSTTVEE239 pKa = 4.56LIGKK243 pKa = 8.6DD244 pKa = 3.06GSMNEE249 pKa = 3.72SRR251 pKa = 11.84ILVNLTVEE259 pKa = 4.39DD260 pKa = 3.98TLAPTIEE267 pKa = 4.74CPSSFDD273 pKa = 3.13ACAGIVNFPDD283 pKa = 3.66AVSNDD288 pKa = 3.26NCTSTVTNTGNVFSGDD304 pKa = 3.44SLTEE308 pKa = 3.97GEE310 pKa = 4.59YY311 pKa = 9.21VTSFEE316 pKa = 4.83AVDD319 pKa = 3.91GSGNVASCTTSFSVNAIPEE338 pKa = 4.04VDD340 pKa = 4.24LGADD344 pKa = 3.43TAVGPGHH351 pKa = 5.93VVSFSVNDD359 pKa = 3.88TVSEE363 pKa = 4.32YY364 pKa = 10.96LWSTGDD370 pKa = 3.37TTRR373 pKa = 11.84SLDD376 pKa = 3.92LKK378 pKa = 10.87VLTDD382 pKa = 3.31TTVWISVTDD391 pKa = 3.93SNSCTGSDD399 pKa = 3.31TATVTTLTSIKK410 pKa = 10.03RR411 pKa = 11.84SKK413 pKa = 9.57KK414 pKa = 7.21TVNVSVYY421 pKa = 9.71PNPTRR426 pKa = 11.84GEE428 pKa = 4.19VKK430 pKa = 10.5VEE432 pKa = 3.7LLGVEE437 pKa = 4.12NLNYY441 pKa = 10.43HH442 pKa = 7.2LIDD445 pKa = 3.31MAGRR449 pKa = 11.84ILQSGNMQNGLNRR462 pKa = 11.84IDD464 pKa = 3.85LSRR467 pKa = 11.84YY468 pKa = 6.07ATSVYY473 pKa = 10.47YY474 pKa = 10.63LQLQNEE480 pKa = 4.3QGEE483 pKa = 4.96GIKK486 pKa = 10.7VMRR489 pKa = 11.84LVKK492 pKa = 10.34QQ493 pKa = 3.31
Molecular weight: 51.8 kDa
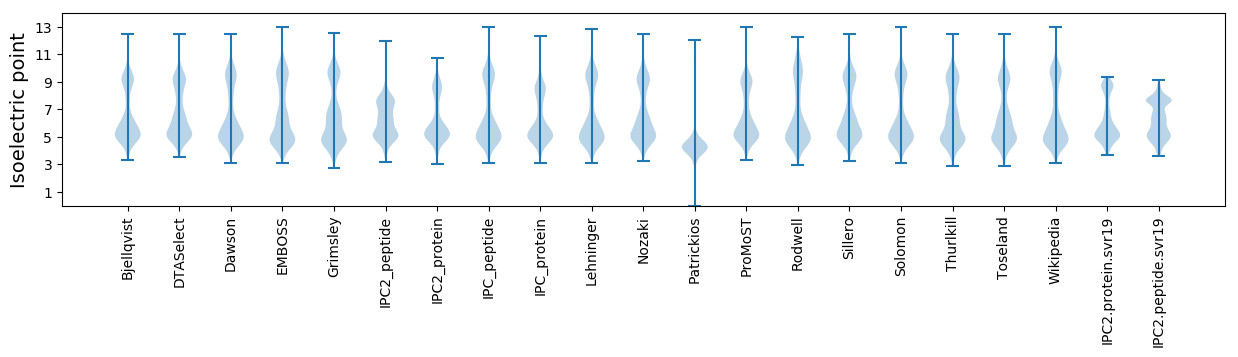
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N6M4S9|A0A6N6M4S9_9FLAO Uncharacterized protein OS=Salibacter halophilus OX=1803916 GN=F3059_07180 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 9.07RR12 pKa = 11.84KK13 pKa = 8.26NKK15 pKa = 9.73HH16 pKa = 4.02GFRR19 pKa = 11.84EE20 pKa = 4.14RR21 pKa = 11.84MSSKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 8.97VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84KK40 pKa = 8.0KK41 pKa = 10.56VSVSSEE47 pKa = 3.8PRR49 pKa = 11.84HH50 pKa = 5.89KK51 pKa = 10.65RR52 pKa = 3.21
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 9.07RR12 pKa = 11.84KK13 pKa = 8.26NKK15 pKa = 9.73HH16 pKa = 4.02GFRR19 pKa = 11.84EE20 pKa = 4.14RR21 pKa = 11.84MSSKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 8.97VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84KK40 pKa = 8.0KK41 pKa = 10.56VSVSSEE47 pKa = 3.8PRR49 pKa = 11.84HH50 pKa = 5.89KK51 pKa = 10.65RR52 pKa = 3.21
Molecular weight: 6.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
957364 |
38 |
3560 |
357.9 |
40.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.06 ± 0.046 | 0.778 ± 0.024 |
6.1 ± 0.035 | 6.993 ± 0.069 |
5.032 ± 0.042 | 6.704 ± 0.047 |
1.81 ± 0.018 | 7.141 ± 0.045 |
7.069 ± 0.078 | 8.791 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.203 ± 0.023 | 5.902 ± 0.053 |
3.571 ± 0.025 | 3.604 ± 0.027 |
3.871 ± 0.047 | 7.146 ± 0.062 |
5.609 ± 0.067 | 6.433 ± 0.035 |
1.11 ± 0.015 | 4.073 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
