
Alteromonas phage vB_AmeM_PT11-V22
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Myoalterovirus; Alteromonas virus PT11-V22
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
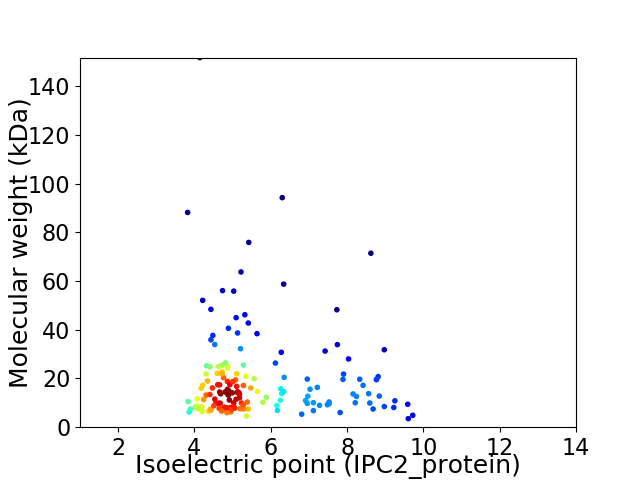
Virtual 2D-PAGE plot for 156 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6C0R1N9|A0A6C0R1N9_9CAUD Thymidylate synthase OS=Alteromonas phage vB_AmeM_PT11-V22 OX=2704031 PE=4 SV=1
MM1 pKa = 7.24GVDD4 pKa = 3.46RR5 pKa = 11.84VAILFRR11 pKa = 11.84DD12 pKa = 3.85TFTEE16 pKa = 4.08SSNTNLIDD24 pKa = 3.47HH25 pKa = 6.6TPEE28 pKa = 3.69QGGPWVYY35 pKa = 10.55EE36 pKa = 3.66AGVPDD41 pKa = 3.36NAAIVRR47 pKa = 11.84SEE49 pKa = 4.62DD50 pKa = 3.49EE51 pKa = 3.92LRR53 pKa = 11.84LNGSIVDD60 pKa = 3.76GFYY63 pKa = 10.95VYY65 pKa = 10.85DD66 pKa = 3.64GTANAGGYY74 pKa = 9.41IATTPASSLVTPEE87 pKa = 4.35KK88 pKa = 9.57YY89 pKa = 10.31CCIRR93 pKa = 11.84LVDD96 pKa = 3.92SNNFFGIEE104 pKa = 3.83FRR106 pKa = 11.84GTGSSGVGFVKK117 pKa = 10.55SVGGVITDD125 pKa = 4.84LYY127 pKa = 11.18SEE129 pKa = 4.42TPEE132 pKa = 4.75AGAVYY137 pKa = 10.78YY138 pKa = 10.96LDD140 pKa = 5.61DD141 pKa = 4.59NGNSLVFKK149 pKa = 9.89KK150 pKa = 10.93DD151 pKa = 3.11NGATTTLYY159 pKa = 10.46TEE161 pKa = 4.34NDD163 pKa = 3.71SEE165 pKa = 5.35FSTEE169 pKa = 3.89TKK171 pKa = 10.27KK172 pKa = 11.15GFVSLGTGNSNPSTYY187 pKa = 10.32YY188 pKa = 10.59EE189 pKa = 4.35SGTLEE194 pKa = 4.36APTGVTINSLSDD206 pKa = 3.31YY207 pKa = 10.06SAKK210 pKa = 10.35PRR212 pKa = 11.84DD213 pKa = 3.89ANGDD217 pKa = 3.33ATFTVSGGCPDD228 pKa = 3.3GTSAIEE234 pKa = 3.84YY235 pKa = 10.39SFDD238 pKa = 3.81EE239 pKa = 4.56ITWLTLDD246 pKa = 4.45ASPSGGAYY254 pKa = 10.32SGTVVVNGQTDD265 pKa = 3.4IFVRR269 pKa = 11.84SADD272 pKa = 3.96DD273 pKa = 3.55NALTASVVGITSAMPIVLWGQSNCDD298 pKa = 2.77GRR300 pKa = 11.84GINNQAVASGSIVPLMYY317 pKa = 10.53KK318 pKa = 10.19SGAVTQMADD327 pKa = 3.32PTGITVDD334 pKa = 3.49SAGSLWPLVVKK345 pKa = 8.99YY346 pKa = 10.14FIDD349 pKa = 3.34QGIPVVMANVAQGGKK364 pKa = 7.43TVAEE368 pKa = 4.49FEE370 pKa = 4.77PSTSLYY376 pKa = 11.04SRR378 pKa = 11.84ISDD381 pKa = 4.01FAANFGSPEE390 pKa = 3.96LAISVIGEE398 pKa = 4.02ADD400 pKa = 3.17SGGTTKK406 pKa = 11.06ADD408 pKa = 3.58FKK410 pKa = 10.93TRR412 pKa = 11.84YY413 pKa = 9.96LNIAQTMNTDD423 pKa = 3.3YY424 pKa = 9.54GTEE427 pKa = 4.03TYY429 pKa = 10.35AVKK432 pKa = 10.57FPVGTATNTTTQTEE446 pKa = 3.66IRR448 pKa = 11.84EE449 pKa = 4.47AYY451 pKa = 10.25DD452 pKa = 3.37EE453 pKa = 5.83LIDD456 pKa = 3.68EE457 pKa = 4.3NAFIKK462 pKa = 10.75FGGDD466 pKa = 3.24LSVIDD471 pKa = 3.77IDD473 pKa = 4.65SGIGHH478 pKa = 7.69DD479 pKa = 4.03SLHH482 pKa = 6.67LKK484 pKa = 8.87TDD486 pKa = 3.19SHH488 pKa = 8.03LSEE491 pKa = 5.72GDD493 pKa = 3.58VIIRR497 pKa = 11.84SALAGIVPDD506 pKa = 3.44VTKK509 pKa = 10.45PVITLLGTPSVVITNGSNYY528 pKa = 9.09TDD530 pKa = 3.72AGATASDD537 pKa = 4.74NIDD540 pKa = 3.17GDD542 pKa = 3.63ITSNIVTVNPVNTNVSATYY561 pKa = 8.89TVTYY565 pKa = 10.59NVTDD569 pKa = 3.73SAGNAADD576 pKa = 3.6EE577 pKa = 4.59VTRR580 pKa = 11.84TVTVQDD586 pKa = 3.83AASDD590 pKa = 3.95NQPPTSNAGADD601 pKa = 3.51QTNIAAGATVTLNGTGSTDD620 pKa = 3.07SDD622 pKa = 3.87GTIVSYY628 pKa = 11.05AWTQTAGDD636 pKa = 3.91IVTLISATSASPTFTAPTTANAQTLTFEE664 pKa = 4.7LTVTDD669 pKa = 5.03DD670 pKa = 4.02GGLTSAPDD678 pKa = 3.58SVDD681 pKa = 3.32VQVNALTLPSVSISAVSQVNTGQSFSPTVTVSGYY715 pKa = 11.33DD716 pKa = 3.41NLLWEE721 pKa = 4.8CTSGQSPSFSSTTAEE736 pKa = 4.27APFITLNEE744 pKa = 4.47GGVHH748 pKa = 5.58TFRR751 pKa = 11.84LTATNISGSVSDD763 pKa = 4.02TFNITGMVQVVEE775 pKa = 4.41SALNLSWEE783 pKa = 4.17GLPDD787 pKa = 3.32GTYY790 pKa = 9.56TVLFIDD796 pKa = 4.37RR797 pKa = 11.84DD798 pKa = 3.69NDD800 pKa = 3.97SVLSRR805 pKa = 11.84SVTSSNEE812 pKa = 3.46AAAVTLPLPVGTKK825 pKa = 9.7VDD827 pKa = 3.97YY828 pKa = 10.39YY829 pKa = 10.3WYY831 pKa = 10.83NEE833 pKa = 4.07TSGDD837 pKa = 3.59AVLNTGATEE846 pKa = 3.85
MM1 pKa = 7.24GVDD4 pKa = 3.46RR5 pKa = 11.84VAILFRR11 pKa = 11.84DD12 pKa = 3.85TFTEE16 pKa = 4.08SSNTNLIDD24 pKa = 3.47HH25 pKa = 6.6TPEE28 pKa = 3.69QGGPWVYY35 pKa = 10.55EE36 pKa = 3.66AGVPDD41 pKa = 3.36NAAIVRR47 pKa = 11.84SEE49 pKa = 4.62DD50 pKa = 3.49EE51 pKa = 3.92LRR53 pKa = 11.84LNGSIVDD60 pKa = 3.76GFYY63 pKa = 10.95VYY65 pKa = 10.85DD66 pKa = 3.64GTANAGGYY74 pKa = 9.41IATTPASSLVTPEE87 pKa = 4.35KK88 pKa = 9.57YY89 pKa = 10.31CCIRR93 pKa = 11.84LVDD96 pKa = 3.92SNNFFGIEE104 pKa = 3.83FRR106 pKa = 11.84GTGSSGVGFVKK117 pKa = 10.55SVGGVITDD125 pKa = 4.84LYY127 pKa = 11.18SEE129 pKa = 4.42TPEE132 pKa = 4.75AGAVYY137 pKa = 10.78YY138 pKa = 10.96LDD140 pKa = 5.61DD141 pKa = 4.59NGNSLVFKK149 pKa = 9.89KK150 pKa = 10.93DD151 pKa = 3.11NGATTTLYY159 pKa = 10.46TEE161 pKa = 4.34NDD163 pKa = 3.71SEE165 pKa = 5.35FSTEE169 pKa = 3.89TKK171 pKa = 10.27KK172 pKa = 11.15GFVSLGTGNSNPSTYY187 pKa = 10.32YY188 pKa = 10.59EE189 pKa = 4.35SGTLEE194 pKa = 4.36APTGVTINSLSDD206 pKa = 3.31YY207 pKa = 10.06SAKK210 pKa = 10.35PRR212 pKa = 11.84DD213 pKa = 3.89ANGDD217 pKa = 3.33ATFTVSGGCPDD228 pKa = 3.3GTSAIEE234 pKa = 3.84YY235 pKa = 10.39SFDD238 pKa = 3.81EE239 pKa = 4.56ITWLTLDD246 pKa = 4.45ASPSGGAYY254 pKa = 10.32SGTVVVNGQTDD265 pKa = 3.4IFVRR269 pKa = 11.84SADD272 pKa = 3.96DD273 pKa = 3.55NALTASVVGITSAMPIVLWGQSNCDD298 pKa = 2.77GRR300 pKa = 11.84GINNQAVASGSIVPLMYY317 pKa = 10.53KK318 pKa = 10.19SGAVTQMADD327 pKa = 3.32PTGITVDD334 pKa = 3.49SAGSLWPLVVKK345 pKa = 8.99YY346 pKa = 10.14FIDD349 pKa = 3.34QGIPVVMANVAQGGKK364 pKa = 7.43TVAEE368 pKa = 4.49FEE370 pKa = 4.77PSTSLYY376 pKa = 11.04SRR378 pKa = 11.84ISDD381 pKa = 4.01FAANFGSPEE390 pKa = 3.96LAISVIGEE398 pKa = 4.02ADD400 pKa = 3.17SGGTTKK406 pKa = 11.06ADD408 pKa = 3.58FKK410 pKa = 10.93TRR412 pKa = 11.84YY413 pKa = 9.96LNIAQTMNTDD423 pKa = 3.3YY424 pKa = 9.54GTEE427 pKa = 4.03TYY429 pKa = 10.35AVKK432 pKa = 10.57FPVGTATNTTTQTEE446 pKa = 3.66IRR448 pKa = 11.84EE449 pKa = 4.47AYY451 pKa = 10.25DD452 pKa = 3.37EE453 pKa = 5.83LIDD456 pKa = 3.68EE457 pKa = 4.3NAFIKK462 pKa = 10.75FGGDD466 pKa = 3.24LSVIDD471 pKa = 3.77IDD473 pKa = 4.65SGIGHH478 pKa = 7.69DD479 pKa = 4.03SLHH482 pKa = 6.67LKK484 pKa = 8.87TDD486 pKa = 3.19SHH488 pKa = 8.03LSEE491 pKa = 5.72GDD493 pKa = 3.58VIIRR497 pKa = 11.84SALAGIVPDD506 pKa = 3.44VTKK509 pKa = 10.45PVITLLGTPSVVITNGSNYY528 pKa = 9.09TDD530 pKa = 3.72AGATASDD537 pKa = 4.74NIDD540 pKa = 3.17GDD542 pKa = 3.63ITSNIVTVNPVNTNVSATYY561 pKa = 8.89TVTYY565 pKa = 10.59NVTDD569 pKa = 3.73SAGNAADD576 pKa = 3.6EE577 pKa = 4.59VTRR580 pKa = 11.84TVTVQDD586 pKa = 3.83AASDD590 pKa = 3.95NQPPTSNAGADD601 pKa = 3.51QTNIAAGATVTLNGTGSTDD620 pKa = 3.07SDD622 pKa = 3.87GTIVSYY628 pKa = 11.05AWTQTAGDD636 pKa = 3.91IVTLISATSASPTFTAPTTANAQTLTFEE664 pKa = 4.7LTVTDD669 pKa = 5.03DD670 pKa = 4.02GGLTSAPDD678 pKa = 3.58SVDD681 pKa = 3.32VQVNALTLPSVSISAVSQVNTGQSFSPTVTVSGYY715 pKa = 11.33DD716 pKa = 3.41NLLWEE721 pKa = 4.8CTSGQSPSFSSTTAEE736 pKa = 4.27APFITLNEE744 pKa = 4.47GGVHH748 pKa = 5.58TFRR751 pKa = 11.84LTATNISGSVSDD763 pKa = 4.02TFNITGMVQVVEE775 pKa = 4.41SALNLSWEE783 pKa = 4.17GLPDD787 pKa = 3.32GTYY790 pKa = 9.56TVLFIDD796 pKa = 4.37RR797 pKa = 11.84DD798 pKa = 3.69NDD800 pKa = 3.97SVLSRR805 pKa = 11.84SVTSSNEE812 pKa = 3.46AAAVTLPLPVGTKK825 pKa = 9.7VDD827 pKa = 3.97YY828 pKa = 10.39YY829 pKa = 10.3WYY831 pKa = 10.83NEE833 pKa = 4.07TSGDD837 pKa = 3.59AVLNTGATEE846 pKa = 3.85
Molecular weight: 88.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6C0R328|A0A6C0R328_9CAUD Uncharacterized protein OS=Alteromonas phage vB_AmeM_PT11-V22 OX=2704031 PE=4 SV=1
MM1 pKa = 7.38HH2 pKa = 7.35FLGRR6 pKa = 11.84SLTTKK11 pKa = 9.6PCSINLRR18 pKa = 11.84VNVLMLPHH26 pKa = 5.99FTISTGSVRR35 pKa = 11.84SVKK38 pKa = 10.09PINLSRR44 pKa = 11.84VLIKK48 pKa = 10.7LEE50 pKa = 3.95YY51 pKa = 8.29VQPFISDD58 pKa = 3.56LTVIAQLVNRR68 pKa = 11.84IVTIFSPLFHH78 pKa = 7.08CGIIANN84 pKa = 4.16
MM1 pKa = 7.38HH2 pKa = 7.35FLGRR6 pKa = 11.84SLTTKK11 pKa = 9.6PCSINLRR18 pKa = 11.84VNVLMLPHH26 pKa = 5.99FTISTGSVRR35 pKa = 11.84SVKK38 pKa = 10.09PINLSRR44 pKa = 11.84VLIKK48 pKa = 10.7LEE50 pKa = 3.95YY51 pKa = 8.29VQPFISDD58 pKa = 3.56LTVIAQLVNRR68 pKa = 11.84IVTIFSPLFHH78 pKa = 7.08CGIIANN84 pKa = 4.16
Molecular weight: 9.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
27604 |
29 |
1430 |
176.9 |
20.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.785 ± 0.302 | 1.33 ± 0.117 |
6.724 ± 0.163 | 8.22 ± 0.256 |
4.148 ± 0.146 | 6.564 ± 0.167 |
1.895 ± 0.135 | 6.245 ± 0.263 |
7.477 ± 0.392 | 8.169 ± 0.201 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.492 ± 0.149 | 5.456 ± 0.25 |
2.945 ± 0.12 | 3.329 ± 0.108 |
3.927 ± 0.148 | 6.51 ± 0.211 |
6.361 ± 0.386 | 6.93 ± 0.205 |
1.337 ± 0.089 | 4.155 ± 0.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
