
Alfalfa mosaic virus (strain 425 / isolate Leiden)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Alfamovirus; Alfalfa mosaic virus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

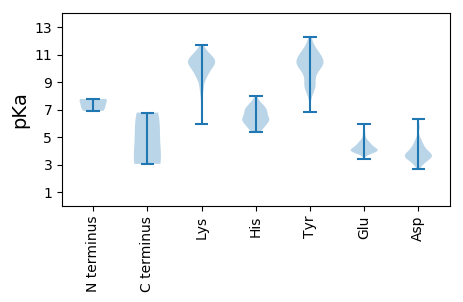
Protein with the lowest isoelectric point:
>sp|P03595|MVP_AMVLE Movement protein OS=Alfalfa mosaic virus (strain 425 / isolate Leiden) OX=12322 GN=ORF3a PE=3 SV=1
MM1 pKa = 6.9FTLLRR6 pKa = 11.84CLGFGVNEE14 pKa = 4.09PTNTSSSEE22 pKa = 4.0YY23 pKa = 8.51VPEE26 pKa = 4.0YY27 pKa = 10.47SVEE30 pKa = 4.17EE31 pKa = 4.0ISNEE35 pKa = 3.91VAEE38 pKa = 4.93LDD40 pKa = 3.83SVDD43 pKa = 5.75PLFQCYY49 pKa = 7.62KK50 pKa = 9.98HH51 pKa = 6.49VFVSLMLVRR60 pKa = 11.84KK61 pKa = 6.15MTQAAEE67 pKa = 4.17DD68 pKa = 3.9FLEE71 pKa = 4.53SFGGEE76 pKa = 3.89FDD78 pKa = 4.27SPCCRR83 pKa = 11.84VYY85 pKa = 10.43RR86 pKa = 11.84LYY88 pKa = 11.1RR89 pKa = 11.84HH90 pKa = 6.23FVNEE94 pKa = 4.03DD95 pKa = 3.56DD96 pKa = 4.17APAWAIPNVVNEE108 pKa = 4.64DD109 pKa = 3.73SYY111 pKa = 12.28DD112 pKa = 3.51DD113 pKa = 3.7YY114 pKa = 11.89AYY116 pKa = 10.93LRR118 pKa = 11.84EE119 pKa = 4.25EE120 pKa = 4.61LDD122 pKa = 4.73AIDD125 pKa = 5.69SSFEE129 pKa = 3.86LLNEE133 pKa = 3.76EE134 pKa = 4.79RR135 pKa = 11.84EE136 pKa = 4.12LSEE139 pKa = 3.79ITDD142 pKa = 3.58RR143 pKa = 11.84LNALRR148 pKa = 11.84FFPVSKK154 pKa = 9.99TEE156 pKa = 4.07ALPVANVQEE165 pKa = 4.27VKK167 pKa = 10.72LISEE171 pKa = 5.21TYY173 pKa = 8.54QLLMTFINYY182 pKa = 9.6SDD184 pKa = 3.94EE185 pKa = 4.75NIPSEE190 pKa = 4.35MPAPLLDD197 pKa = 4.06EE198 pKa = 5.84LGMLPEE204 pKa = 4.36EE205 pKa = 4.65LGPLNEE211 pKa = 4.71IEE213 pKa = 4.97DD214 pKa = 4.41IKK216 pKa = 11.04PVAAPITLLSEE227 pKa = 4.36FRR229 pKa = 11.84ASDD232 pKa = 3.31NAKK235 pKa = 9.86PLDD238 pKa = 3.7IVEE241 pKa = 5.39IIPDD245 pKa = 3.67VSPTKK250 pKa = 10.07PYY252 pKa = 10.53EE253 pKa = 3.99AVISGNDD260 pKa = 2.84WMTLGRR266 pKa = 11.84IIPTTPVPTIRR277 pKa = 11.84DD278 pKa = 3.52VFFSGLSRR286 pKa = 11.84HH287 pKa = 6.0GSPEE291 pKa = 4.06VIQNALDD298 pKa = 3.52EE299 pKa = 4.7FLPLHH304 pKa = 6.44HH305 pKa = 7.23SIDD308 pKa = 3.44DD309 pKa = 3.77KK310 pKa = 11.71YY311 pKa = 9.29FQEE314 pKa = 4.24WVEE317 pKa = 4.15TSDD320 pKa = 4.5KK321 pKa = 11.33SLDD324 pKa = 3.54VDD326 pKa = 3.78PCRR329 pKa = 11.84IDD331 pKa = 4.89LSVFNNWQSSEE342 pKa = 3.77NCYY345 pKa = 10.13EE346 pKa = 3.88PRR348 pKa = 11.84FKK350 pKa = 10.39TGALSTRR357 pKa = 11.84KK358 pKa = 8.41GTQTEE363 pKa = 3.84ALLAIKK369 pKa = 9.37KK370 pKa = 10.41RR371 pKa = 11.84NMNVPNLGQIYY382 pKa = 10.06DD383 pKa = 3.92VNSVANSVVNKK394 pKa = 10.41LLTTVIDD401 pKa = 3.97PDD403 pKa = 4.33KK404 pKa = 11.43LCMFPDD410 pKa = 5.76FISEE414 pKa = 4.42GEE416 pKa = 3.85VSYY419 pKa = 10.54FQDD422 pKa = 3.77YY423 pKa = 10.88IVGKK427 pKa = 10.24NPDD430 pKa = 3.78PEE432 pKa = 5.45LYY434 pKa = 10.37SDD436 pKa = 3.93PLGVRR441 pKa = 11.84SIDD444 pKa = 3.73SYY446 pKa = 11.36KK447 pKa = 11.12HH448 pKa = 5.52MIKK451 pKa = 10.34SVLKK455 pKa = 9.54PVEE458 pKa = 4.59DD459 pKa = 3.68NSLHH463 pKa = 6.86LEE465 pKa = 4.08RR466 pKa = 11.84PMPATITYY474 pKa = 8.77HH475 pKa = 7.14DD476 pKa = 4.57KK477 pKa = 11.09DD478 pKa = 3.48IVMSSSPIFLAAAARR493 pKa = 11.84LMLILRR499 pKa = 11.84DD500 pKa = 4.09KK501 pKa = 10.37ITIPSGKK508 pKa = 9.15FHH510 pKa = 6.67QLFSIDD516 pKa = 4.43AEE518 pKa = 4.06AFDD521 pKa = 4.73ASFHH525 pKa = 5.89FKK527 pKa = 10.59EE528 pKa = 3.9IDD530 pKa = 3.31FSKK533 pKa = 10.58FDD535 pKa = 3.28KK536 pKa = 10.89SQNEE540 pKa = 4.02LHH542 pKa = 6.83HH543 pKa = 6.82LIQEE547 pKa = 4.37RR548 pKa = 11.84FLKK551 pKa = 10.83YY552 pKa = 10.44LGIPNEE558 pKa = 4.48FLTLWFNAHH567 pKa = 5.81RR568 pKa = 11.84KK569 pKa = 9.26SRR571 pKa = 11.84ISDD574 pKa = 3.44SKK576 pKa = 11.08NGVFFNVDD584 pKa = 3.72FQRR587 pKa = 11.84RR588 pKa = 11.84TGDD591 pKa = 3.07ALTYY595 pKa = 10.34LGNTIVTLACLCHH608 pKa = 7.66VYY610 pKa = 11.25DD611 pKa = 5.97LMDD614 pKa = 4.9PNVKK618 pKa = 9.9FVVASGDD625 pKa = 3.45DD626 pKa = 3.72SLIGTVEE633 pKa = 3.98EE634 pKa = 4.55LPRR637 pKa = 11.84DD638 pKa = 3.62QEE640 pKa = 4.42FLFTTLFNLEE650 pKa = 3.95AKK652 pKa = 10.2FPHH655 pKa = 5.97NQPFICSKK663 pKa = 10.61FLITMPTTSGGKK675 pKa = 9.38VVLPIPNPLKK685 pKa = 10.72LLIRR689 pKa = 11.84LGSKK693 pKa = 9.63KK694 pKa = 10.5VNADD698 pKa = 4.42IFDD701 pKa = 3.33EE702 pKa = 5.39WYY704 pKa = 10.56QSWIDD709 pKa = 3.71IIGGFNDD716 pKa = 3.21HH717 pKa = 6.83HH718 pKa = 7.25VIRR721 pKa = 11.84CVAAMTAHH729 pKa = 7.23RR730 pKa = 11.84YY731 pKa = 8.54LRR733 pKa = 11.84RR734 pKa = 11.84PSLYY738 pKa = 10.67LEE740 pKa = 4.62AALEE744 pKa = 4.16SLGKK748 pKa = 10.13IFAGKK753 pKa = 7.12TLCKK757 pKa = 10.16EE758 pKa = 3.93CLFNEE763 pKa = 4.07KK764 pKa = 10.31HH765 pKa = 5.65EE766 pKa = 5.19SNVKK770 pKa = 9.5IKK772 pKa = 9.98PRR774 pKa = 11.84RR775 pKa = 11.84VKK777 pKa = 10.42KK778 pKa = 9.65SHH780 pKa = 6.2SDD782 pKa = 2.85ARR784 pKa = 11.84SRR786 pKa = 11.84ARR788 pKa = 11.84RR789 pKa = 11.84AA790 pKa = 3.05
MM1 pKa = 6.9FTLLRR6 pKa = 11.84CLGFGVNEE14 pKa = 4.09PTNTSSSEE22 pKa = 4.0YY23 pKa = 8.51VPEE26 pKa = 4.0YY27 pKa = 10.47SVEE30 pKa = 4.17EE31 pKa = 4.0ISNEE35 pKa = 3.91VAEE38 pKa = 4.93LDD40 pKa = 3.83SVDD43 pKa = 5.75PLFQCYY49 pKa = 7.62KK50 pKa = 9.98HH51 pKa = 6.49VFVSLMLVRR60 pKa = 11.84KK61 pKa = 6.15MTQAAEE67 pKa = 4.17DD68 pKa = 3.9FLEE71 pKa = 4.53SFGGEE76 pKa = 3.89FDD78 pKa = 4.27SPCCRR83 pKa = 11.84VYY85 pKa = 10.43RR86 pKa = 11.84LYY88 pKa = 11.1RR89 pKa = 11.84HH90 pKa = 6.23FVNEE94 pKa = 4.03DD95 pKa = 3.56DD96 pKa = 4.17APAWAIPNVVNEE108 pKa = 4.64DD109 pKa = 3.73SYY111 pKa = 12.28DD112 pKa = 3.51DD113 pKa = 3.7YY114 pKa = 11.89AYY116 pKa = 10.93LRR118 pKa = 11.84EE119 pKa = 4.25EE120 pKa = 4.61LDD122 pKa = 4.73AIDD125 pKa = 5.69SSFEE129 pKa = 3.86LLNEE133 pKa = 3.76EE134 pKa = 4.79RR135 pKa = 11.84EE136 pKa = 4.12LSEE139 pKa = 3.79ITDD142 pKa = 3.58RR143 pKa = 11.84LNALRR148 pKa = 11.84FFPVSKK154 pKa = 9.99TEE156 pKa = 4.07ALPVANVQEE165 pKa = 4.27VKK167 pKa = 10.72LISEE171 pKa = 5.21TYY173 pKa = 8.54QLLMTFINYY182 pKa = 9.6SDD184 pKa = 3.94EE185 pKa = 4.75NIPSEE190 pKa = 4.35MPAPLLDD197 pKa = 4.06EE198 pKa = 5.84LGMLPEE204 pKa = 4.36EE205 pKa = 4.65LGPLNEE211 pKa = 4.71IEE213 pKa = 4.97DD214 pKa = 4.41IKK216 pKa = 11.04PVAAPITLLSEE227 pKa = 4.36FRR229 pKa = 11.84ASDD232 pKa = 3.31NAKK235 pKa = 9.86PLDD238 pKa = 3.7IVEE241 pKa = 5.39IIPDD245 pKa = 3.67VSPTKK250 pKa = 10.07PYY252 pKa = 10.53EE253 pKa = 3.99AVISGNDD260 pKa = 2.84WMTLGRR266 pKa = 11.84IIPTTPVPTIRR277 pKa = 11.84DD278 pKa = 3.52VFFSGLSRR286 pKa = 11.84HH287 pKa = 6.0GSPEE291 pKa = 4.06VIQNALDD298 pKa = 3.52EE299 pKa = 4.7FLPLHH304 pKa = 6.44HH305 pKa = 7.23SIDD308 pKa = 3.44DD309 pKa = 3.77KK310 pKa = 11.71YY311 pKa = 9.29FQEE314 pKa = 4.24WVEE317 pKa = 4.15TSDD320 pKa = 4.5KK321 pKa = 11.33SLDD324 pKa = 3.54VDD326 pKa = 3.78PCRR329 pKa = 11.84IDD331 pKa = 4.89LSVFNNWQSSEE342 pKa = 3.77NCYY345 pKa = 10.13EE346 pKa = 3.88PRR348 pKa = 11.84FKK350 pKa = 10.39TGALSTRR357 pKa = 11.84KK358 pKa = 8.41GTQTEE363 pKa = 3.84ALLAIKK369 pKa = 9.37KK370 pKa = 10.41RR371 pKa = 11.84NMNVPNLGQIYY382 pKa = 10.06DD383 pKa = 3.92VNSVANSVVNKK394 pKa = 10.41LLTTVIDD401 pKa = 3.97PDD403 pKa = 4.33KK404 pKa = 11.43LCMFPDD410 pKa = 5.76FISEE414 pKa = 4.42GEE416 pKa = 3.85VSYY419 pKa = 10.54FQDD422 pKa = 3.77YY423 pKa = 10.88IVGKK427 pKa = 10.24NPDD430 pKa = 3.78PEE432 pKa = 5.45LYY434 pKa = 10.37SDD436 pKa = 3.93PLGVRR441 pKa = 11.84SIDD444 pKa = 3.73SYY446 pKa = 11.36KK447 pKa = 11.12HH448 pKa = 5.52MIKK451 pKa = 10.34SVLKK455 pKa = 9.54PVEE458 pKa = 4.59DD459 pKa = 3.68NSLHH463 pKa = 6.86LEE465 pKa = 4.08RR466 pKa = 11.84PMPATITYY474 pKa = 8.77HH475 pKa = 7.14DD476 pKa = 4.57KK477 pKa = 11.09DD478 pKa = 3.48IVMSSSPIFLAAAARR493 pKa = 11.84LMLILRR499 pKa = 11.84DD500 pKa = 4.09KK501 pKa = 10.37ITIPSGKK508 pKa = 9.15FHH510 pKa = 6.67QLFSIDD516 pKa = 4.43AEE518 pKa = 4.06AFDD521 pKa = 4.73ASFHH525 pKa = 5.89FKK527 pKa = 10.59EE528 pKa = 3.9IDD530 pKa = 3.31FSKK533 pKa = 10.58FDD535 pKa = 3.28KK536 pKa = 10.89SQNEE540 pKa = 4.02LHH542 pKa = 6.83HH543 pKa = 6.82LIQEE547 pKa = 4.37RR548 pKa = 11.84FLKK551 pKa = 10.83YY552 pKa = 10.44LGIPNEE558 pKa = 4.48FLTLWFNAHH567 pKa = 5.81RR568 pKa = 11.84KK569 pKa = 9.26SRR571 pKa = 11.84ISDD574 pKa = 3.44SKK576 pKa = 11.08NGVFFNVDD584 pKa = 3.72FQRR587 pKa = 11.84RR588 pKa = 11.84TGDD591 pKa = 3.07ALTYY595 pKa = 10.34LGNTIVTLACLCHH608 pKa = 7.66VYY610 pKa = 11.25DD611 pKa = 5.97LMDD614 pKa = 4.9PNVKK618 pKa = 9.9FVVASGDD625 pKa = 3.45DD626 pKa = 3.72SLIGTVEE633 pKa = 3.98EE634 pKa = 4.55LPRR637 pKa = 11.84DD638 pKa = 3.62QEE640 pKa = 4.42FLFTTLFNLEE650 pKa = 3.95AKK652 pKa = 10.2FPHH655 pKa = 5.97NQPFICSKK663 pKa = 10.61FLITMPTTSGGKK675 pKa = 9.38VVLPIPNPLKK685 pKa = 10.72LLIRR689 pKa = 11.84LGSKK693 pKa = 9.63KK694 pKa = 10.5VNADD698 pKa = 4.42IFDD701 pKa = 3.33EE702 pKa = 5.39WYY704 pKa = 10.56QSWIDD709 pKa = 3.71IIGGFNDD716 pKa = 3.21HH717 pKa = 6.83HH718 pKa = 7.25VIRR721 pKa = 11.84CVAAMTAHH729 pKa = 7.23RR730 pKa = 11.84YY731 pKa = 8.54LRR733 pKa = 11.84RR734 pKa = 11.84PSLYY738 pKa = 10.67LEE740 pKa = 4.62AALEE744 pKa = 4.16SLGKK748 pKa = 10.13IFAGKK753 pKa = 7.12TLCKK757 pKa = 10.16EE758 pKa = 3.93CLFNEE763 pKa = 4.07KK764 pKa = 10.31HH765 pKa = 5.65EE766 pKa = 5.19SNVKK770 pKa = 9.5IKK772 pKa = 9.98PRR774 pKa = 11.84RR775 pKa = 11.84VKK777 pKa = 10.42KK778 pKa = 9.65SHH780 pKa = 6.2SDD782 pKa = 2.85ARR784 pKa = 11.84SRR786 pKa = 11.84ARR788 pKa = 11.84RR789 pKa = 11.84AA790 pKa = 3.05
Molecular weight: 89.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03593|RDRP_AMVLE RNA-directed RNA polymerase 2a OS=Alfalfa mosaic virus (strain 425 / isolate Leiden) OX=12322 GN=ORF2a PE=3 SV=2
MM1 pKa = 7.77SSSQKK6 pKa = 10.31KK7 pKa = 10.38AGGKK11 pKa = 9.36AGKK14 pKa = 6.82PTKK17 pKa = 10.18RR18 pKa = 11.84SQNYY22 pKa = 8.54AALRR26 pKa = 11.84KK27 pKa = 9.29AQLPKK32 pKa = 10.5PPALKK37 pKa = 10.71VPVVKK42 pKa = 8.86PTNTILPQTGCVWQSLGTPLSLSSFNGLGARR73 pKa = 11.84FLYY76 pKa = 10.8SFLKK80 pKa = 10.82DD81 pKa = 3.48FVGPRR86 pKa = 11.84ILEE89 pKa = 3.81EE90 pKa = 3.9DD91 pKa = 4.35LIYY94 pKa = 11.15RR95 pKa = 11.84MVFSITPSHH104 pKa = 6.87AGTFCLTDD112 pKa = 3.62DD113 pKa = 3.77VTTEE117 pKa = 3.97DD118 pKa = 3.79GRR120 pKa = 11.84AVAHH124 pKa = 6.19GNPMQEE130 pKa = 4.26FPHH133 pKa = 6.38GAFHH137 pKa = 7.23ANEE140 pKa = 4.02KK141 pKa = 10.73FGFEE145 pKa = 4.39LVFTAPTHH153 pKa = 7.53AGMQNQNFKK162 pKa = 10.65HH163 pKa = 6.06SYY165 pKa = 10.11AVALCLDD172 pKa = 4.68FDD174 pKa = 4.28AQPEE178 pKa = 4.57GSKK181 pKa = 10.81NPSFRR186 pKa = 11.84FNEE189 pKa = 3.79VWVEE193 pKa = 4.28RR194 pKa = 11.84KK195 pKa = 9.65AFPRR199 pKa = 11.84AGPLRR204 pKa = 11.84SLITVGLFDD213 pKa = 5.08EE214 pKa = 5.92ADD216 pKa = 3.68DD217 pKa = 4.83LDD219 pKa = 3.91RR220 pKa = 11.84HH221 pKa = 5.68
MM1 pKa = 7.77SSSQKK6 pKa = 10.31KK7 pKa = 10.38AGGKK11 pKa = 9.36AGKK14 pKa = 6.82PTKK17 pKa = 10.18RR18 pKa = 11.84SQNYY22 pKa = 8.54AALRR26 pKa = 11.84KK27 pKa = 9.29AQLPKK32 pKa = 10.5PPALKK37 pKa = 10.71VPVVKK42 pKa = 8.86PTNTILPQTGCVWQSLGTPLSLSSFNGLGARR73 pKa = 11.84FLYY76 pKa = 10.8SFLKK80 pKa = 10.82DD81 pKa = 3.48FVGPRR86 pKa = 11.84ILEE89 pKa = 3.81EE90 pKa = 3.9DD91 pKa = 4.35LIYY94 pKa = 11.15RR95 pKa = 11.84MVFSITPSHH104 pKa = 6.87AGTFCLTDD112 pKa = 3.62DD113 pKa = 3.77VTTEE117 pKa = 3.97DD118 pKa = 3.79GRR120 pKa = 11.84AVAHH124 pKa = 6.19GNPMQEE130 pKa = 4.26FPHH133 pKa = 6.38GAFHH137 pKa = 7.23ANEE140 pKa = 4.02KK141 pKa = 10.73FGFEE145 pKa = 4.39LVFTAPTHH153 pKa = 7.53AGMQNQNFKK162 pKa = 10.65HH163 pKa = 6.06SYY165 pKa = 10.11AVALCLDD172 pKa = 4.68FDD174 pKa = 4.28AQPEE178 pKa = 4.57GSKK181 pKa = 10.81NPSFRR186 pKa = 11.84FNEE189 pKa = 3.79VWVEE193 pKa = 4.28RR194 pKa = 11.84KK195 pKa = 9.65AFPRR199 pKa = 11.84AGPLRR204 pKa = 11.84SLITVGLFDD213 pKa = 5.08EE214 pKa = 5.92ADD216 pKa = 3.68DD217 pKa = 4.83LDD219 pKa = 3.91RR220 pKa = 11.84HH221 pKa = 5.68
Molecular weight: 24.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2437 |
221 |
1126 |
609.3 |
68.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.427 ± 0.707 | 1.805 ± 0.325 |
6.524 ± 0.517 | 5.745 ± 0.689 |
4.924 ± 0.671 | 5.211 ± 0.755 |
2.831 ± 0.228 | 5.745 ± 0.674 |
6.278 ± 0.392 | 9.561 ± 0.549 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.929 ± 0.242 | 4.391 ± 0.267 |
5.252 ± 0.73 | 2.544 ± 0.312 |
5.129 ± 0.395 | 8.453 ± 0.521 |
5.622 ± 0.374 | 6.648 ± 0.134 |
0.78 ± 0.091 | 3.201 ± 0.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
