
Hanseniaspora uvarum (Yeast) (Kloeckera apiculata)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
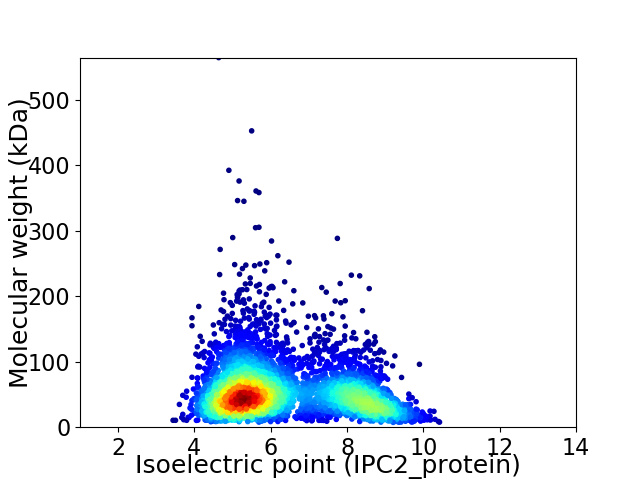
Virtual 2D-PAGE plot for 4059 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E5S1Z9|A0A1E5S1Z9_HANUV Ceramide synthase subunit LIP1 OS=Hanseniaspora uvarum OX=29833 GN=AWRI3580_g76 PE=4 SV=1
MM1 pKa = 7.59QYY3 pKa = 7.88TTITSIFLLTISLLTQSTSAIGTLGINVASSGVDD37 pKa = 3.5GNCRR41 pKa = 11.84DD42 pKa = 3.53SDD44 pKa = 3.72SWSEE48 pKa = 4.07ALEE51 pKa = 4.17KK52 pKa = 10.92ASAFTNYY59 pKa = 8.96IKK61 pKa = 10.0TYY63 pKa = 10.62AVSDD67 pKa = 4.0CNMLEE72 pKa = 4.27NIVPALNSNTNMKK85 pKa = 9.47VWMGVWEE92 pKa = 4.04VDD94 pKa = 3.35EE95 pKa = 4.64AHH97 pKa = 7.04FDD99 pKa = 3.72AEE101 pKa = 4.39KK102 pKa = 10.96DD103 pKa = 3.38ALTTYY108 pKa = 10.44LPSISKK114 pKa = 10.62DD115 pKa = 3.43SIAGISVGSEE125 pKa = 3.03ALYY128 pKa = 10.84RR129 pKa = 11.84GDD131 pKa = 4.37LTPQALASKK140 pKa = 9.91ISEE143 pKa = 4.13IKK145 pKa = 10.68SLLSDD150 pKa = 3.27ITDD153 pKa = 3.46KK154 pKa = 11.5DD155 pKa = 3.85GNSYY159 pKa = 11.03SGVSVGTVDD168 pKa = 4.59SWNIWVNGTNAPAIQAADD186 pKa = 3.9YY187 pKa = 11.22IMVNAFPFWQYY198 pKa = 7.92QTANNQSHH206 pKa = 6.35SLVDD210 pKa = 4.17DD211 pKa = 3.7VMQAVKK217 pKa = 10.51AIQEE221 pKa = 4.21AKK223 pKa = 9.59GTDD226 pKa = 3.3DD227 pKa = 4.77FYY229 pKa = 11.13ISVGEE234 pKa = 4.27TGAPTGGNEE243 pKa = 4.19LTSGQAPLTVDD254 pKa = 3.1NASDD258 pKa = 4.12LFQEE262 pKa = 5.73GICTLLAYY270 pKa = 10.29GIDD273 pKa = 3.66VNIFEE278 pKa = 5.22LQDD281 pKa = 3.54EE282 pKa = 4.58PTKK285 pKa = 10.67PLATADD291 pKa = 4.55DD292 pKa = 4.87GSSSDD297 pKa = 4.1VEE299 pKa = 4.44RR300 pKa = 11.84HH301 pKa = 4.36WGVYY305 pKa = 8.53DD306 pKa = 3.19TDD308 pKa = 4.08YY309 pKa = 11.28NLKK312 pKa = 10.61YY313 pKa = 10.76SLTCSYY319 pKa = 11.2GSNN322 pKa = 3.33
MM1 pKa = 7.59QYY3 pKa = 7.88TTITSIFLLTISLLTQSTSAIGTLGINVASSGVDD37 pKa = 3.5GNCRR41 pKa = 11.84DD42 pKa = 3.53SDD44 pKa = 3.72SWSEE48 pKa = 4.07ALEE51 pKa = 4.17KK52 pKa = 10.92ASAFTNYY59 pKa = 8.96IKK61 pKa = 10.0TYY63 pKa = 10.62AVSDD67 pKa = 4.0CNMLEE72 pKa = 4.27NIVPALNSNTNMKK85 pKa = 9.47VWMGVWEE92 pKa = 4.04VDD94 pKa = 3.35EE95 pKa = 4.64AHH97 pKa = 7.04FDD99 pKa = 3.72AEE101 pKa = 4.39KK102 pKa = 10.96DD103 pKa = 3.38ALTTYY108 pKa = 10.44LPSISKK114 pKa = 10.62DD115 pKa = 3.43SIAGISVGSEE125 pKa = 3.03ALYY128 pKa = 10.84RR129 pKa = 11.84GDD131 pKa = 4.37LTPQALASKK140 pKa = 9.91ISEE143 pKa = 4.13IKK145 pKa = 10.68SLLSDD150 pKa = 3.27ITDD153 pKa = 3.46KK154 pKa = 11.5DD155 pKa = 3.85GNSYY159 pKa = 11.03SGVSVGTVDD168 pKa = 4.59SWNIWVNGTNAPAIQAADD186 pKa = 3.9YY187 pKa = 11.22IMVNAFPFWQYY198 pKa = 7.92QTANNQSHH206 pKa = 6.35SLVDD210 pKa = 4.17DD211 pKa = 3.7VMQAVKK217 pKa = 10.51AIQEE221 pKa = 4.21AKK223 pKa = 9.59GTDD226 pKa = 3.3DD227 pKa = 4.77FYY229 pKa = 11.13ISVGEE234 pKa = 4.27TGAPTGGNEE243 pKa = 4.19LTSGQAPLTVDD254 pKa = 3.1NASDD258 pKa = 4.12LFQEE262 pKa = 5.73GICTLLAYY270 pKa = 10.29GIDD273 pKa = 3.66VNIFEE278 pKa = 5.22LQDD281 pKa = 3.54EE282 pKa = 4.58PTKK285 pKa = 10.67PLATADD291 pKa = 4.55DD292 pKa = 4.87GSSSDD297 pKa = 4.1VEE299 pKa = 4.44RR300 pKa = 11.84HH301 pKa = 4.36WGVYY305 pKa = 8.53DD306 pKa = 3.19TDD308 pKa = 4.08YY309 pKa = 11.28NLKK312 pKa = 10.61YY313 pKa = 10.76SLTCSYY319 pKa = 11.2GSNN322 pKa = 3.33
Molecular weight: 34.66 kDa
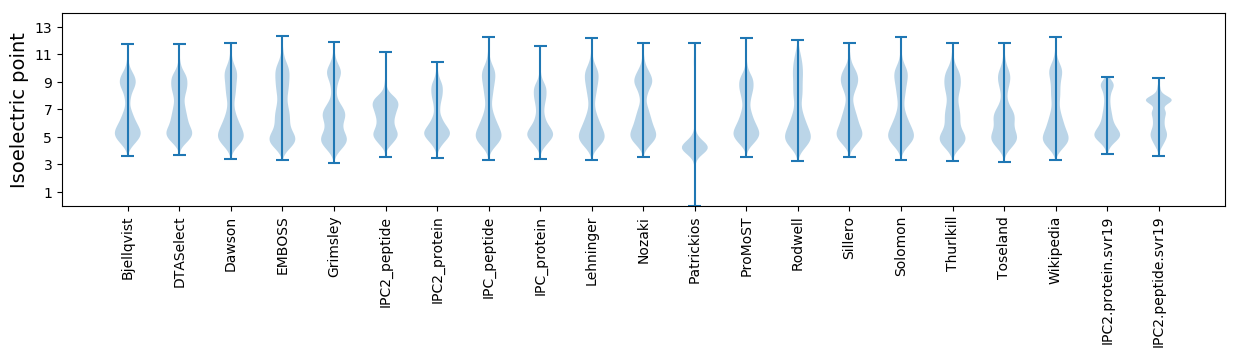
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E5RJZ3|A0A1E5RJZ3_HANUV Cell division control protein 53 OS=Hanseniaspora uvarum OX=29833 GN=AWRI3580_g2889 PE=3 SV=1
MM1 pKa = 7.52DD2 pKa = 5.56TIRR5 pKa = 11.84TGITGKK11 pKa = 10.68YY12 pKa = 7.27GVRR15 pKa = 11.84YY16 pKa = 9.34GSSLRR21 pKa = 11.84RR22 pKa = 11.84QVKK25 pKa = 9.69KK26 pKa = 11.14LEE28 pKa = 4.21VQQHH32 pKa = 4.66SRR34 pKa = 11.84YY35 pKa = 10.26DD36 pKa = 3.35CSFCGKK42 pKa = 10.23KK43 pKa = 9.77SVKK46 pKa = 10.2RR47 pKa = 11.84GATGIWNCGSCKK59 pKa = 9.43KK60 pKa = 8.82TVAGGAYY67 pKa = 8.32TVSTAAAATVRR78 pKa = 11.84STIRR82 pKa = 11.84RR83 pKa = 11.84LRR85 pKa = 11.84DD86 pKa = 3.24MVEE89 pKa = 3.53AA90 pKa = 4.79
MM1 pKa = 7.52DD2 pKa = 5.56TIRR5 pKa = 11.84TGITGKK11 pKa = 10.68YY12 pKa = 7.27GVRR15 pKa = 11.84YY16 pKa = 9.34GSSLRR21 pKa = 11.84RR22 pKa = 11.84QVKK25 pKa = 9.69KK26 pKa = 11.14LEE28 pKa = 4.21VQQHH32 pKa = 4.66SRR34 pKa = 11.84YY35 pKa = 10.26DD36 pKa = 3.35CSFCGKK42 pKa = 10.23KK43 pKa = 9.77SVKK46 pKa = 10.2RR47 pKa = 11.84GATGIWNCGSCKK59 pKa = 9.43KK60 pKa = 8.82TVAGGAYY67 pKa = 8.32TVSTAAAATVRR78 pKa = 11.84STIRR82 pKa = 11.84RR83 pKa = 11.84LRR85 pKa = 11.84DD86 pKa = 3.24MVEE89 pKa = 3.53AA90 pKa = 4.79
Molecular weight: 9.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2125948 |
66 |
4955 |
523.8 |
59.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.36 ± 0.036 | 1.123 ± 0.011 |
6.269 ± 0.03 | 6.933 ± 0.035 |
4.801 ± 0.025 | 4.332 ± 0.041 |
1.799 ± 0.012 | 7.475 ± 0.033 |
8.957 ± 0.044 | 9.61 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.126 ± 0.015 | 7.721 ± 0.048 |
3.361 ± 0.025 | 3.674 ± 0.022 |
3.08 ± 0.021 | 8.31 ± 0.052 |
5.523 ± 0.042 | 5.354 ± 0.023 |
0.822 ± 0.01 | 3.864 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
