
Microbacterium paludicola
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
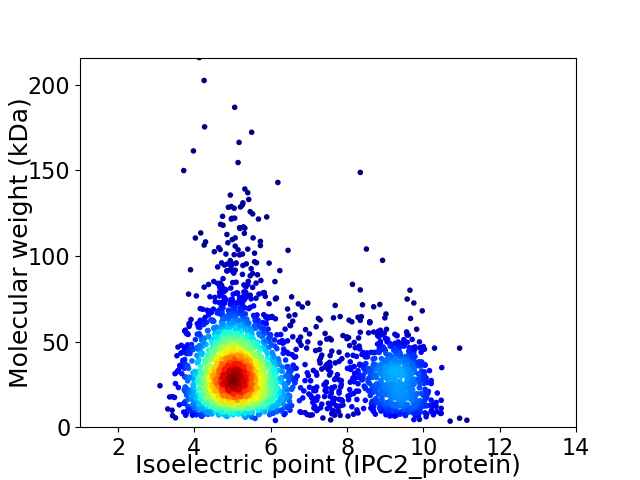
Virtual 2D-PAGE plot for 3006 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9FSB3|A0A4Y9FSB3_9MICO Aminodeoxychorismate lyase OS=Microbacterium paludicola OX=300019 GN=E4U02_12240 PE=4 SV=1
MM1 pKa = 6.84VRR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 10.0LIGALALTGAAALTLAGCATGGGGTATEE34 pKa = 4.92DD35 pKa = 3.72PSAGPAEE42 pKa = 4.43PGGDD46 pKa = 3.73LSLKK50 pKa = 10.1IATALPVTGSLAFLGPPEE68 pKa = 4.08IQGVNYY74 pKa = 9.83AQSLINEE81 pKa = 4.39YY82 pKa = 9.7TEE84 pKa = 4.19EE85 pKa = 4.15TGLEE89 pKa = 4.08LDD91 pKa = 4.23VVHH94 pKa = 7.05GDD96 pKa = 5.45SGDD99 pKa = 4.01LDD101 pKa = 3.69NKK103 pKa = 10.28KK104 pKa = 10.55YY105 pKa = 10.36EE106 pKa = 4.19SEE108 pKa = 3.93IPRR111 pKa = 11.84LLSEE115 pKa = 5.09DD116 pKa = 3.25PAAIIGAASSGVSLQFIDD134 pKa = 3.62QVVGAGVIQFSPANTSDD151 pKa = 3.9AFTTYY156 pKa = 10.99DD157 pKa = 3.97DD158 pKa = 3.37NGLYY162 pKa = 10.42FRR164 pKa = 11.84TAPSDD169 pKa = 3.6VLQGEE174 pKa = 4.35VHH176 pKa = 6.59GNFLGEE182 pKa = 4.92LGHH185 pKa = 5.7QTLGLIVLNDD195 pKa = 3.6AYY197 pKa = 10.34GTGLAKK203 pKa = 10.77YY204 pKa = 8.47ITEE207 pKa = 4.17AFEE210 pKa = 4.32GAGGEE215 pKa = 4.5VVASEE220 pKa = 4.39TFNPGDD226 pKa = 3.6TSFDD230 pKa = 3.61SQIAAVLAEE239 pKa = 5.17DD240 pKa = 4.36PDD242 pKa = 5.98AITVISFDD250 pKa = 3.56EE251 pKa = 4.73AKK253 pKa = 10.12TILPSLIEE261 pKa = 4.12KK262 pKa = 10.78GFDD265 pKa = 3.25AEE267 pKa = 4.85NLFLVDD273 pKa = 4.63GNMAAYY279 pKa = 10.28DD280 pKa = 3.75KK281 pKa = 10.96DD282 pKa = 4.19FPAGLLEE289 pKa = 4.51GAKK292 pKa = 8.77GTYY295 pKa = 9.39PGPTPDD301 pKa = 3.18QVQEE305 pKa = 4.19FVDD308 pKa = 3.84GLNAFVEE315 pKa = 4.64EE316 pKa = 4.76SGTEE320 pKa = 3.8ALSDD324 pKa = 3.39YY325 pKa = 10.26TYY327 pKa = 11.26APEE330 pKa = 5.11SFDD333 pKa = 3.71AVNLIALASLAAQSTEE349 pKa = 3.83PADD352 pKa = 3.39IAEE355 pKa = 4.49KK356 pKa = 10.35LVEE359 pKa = 4.02VSGGSGEE366 pKa = 4.46GEE368 pKa = 4.2KK369 pKa = 9.9CTSFADD375 pKa = 3.8CADD378 pKa = 4.17IILGGGVADD387 pKa = 3.97YY388 pKa = 10.9DD389 pKa = 4.25GASSPVTFDD398 pKa = 3.31EE399 pKa = 4.97VGDD402 pKa = 3.89PTEE405 pKa = 4.02GMINIYY411 pKa = 10.38EE412 pKa = 4.03YY413 pKa = 11.22GADD416 pKa = 3.43NKK418 pKa = 10.19YY419 pKa = 10.76VPYY422 pKa = 10.56EE423 pKa = 3.99GG424 pKa = 4.4
MM1 pKa = 6.84VRR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 10.0LIGALALTGAAALTLAGCATGGGGTATEE34 pKa = 4.92DD35 pKa = 3.72PSAGPAEE42 pKa = 4.43PGGDD46 pKa = 3.73LSLKK50 pKa = 10.1IATALPVTGSLAFLGPPEE68 pKa = 4.08IQGVNYY74 pKa = 9.83AQSLINEE81 pKa = 4.39YY82 pKa = 9.7TEE84 pKa = 4.19EE85 pKa = 4.15TGLEE89 pKa = 4.08LDD91 pKa = 4.23VVHH94 pKa = 7.05GDD96 pKa = 5.45SGDD99 pKa = 4.01LDD101 pKa = 3.69NKK103 pKa = 10.28KK104 pKa = 10.55YY105 pKa = 10.36EE106 pKa = 4.19SEE108 pKa = 3.93IPRR111 pKa = 11.84LLSEE115 pKa = 5.09DD116 pKa = 3.25PAAIIGAASSGVSLQFIDD134 pKa = 3.62QVVGAGVIQFSPANTSDD151 pKa = 3.9AFTTYY156 pKa = 10.99DD157 pKa = 3.97DD158 pKa = 3.37NGLYY162 pKa = 10.42FRR164 pKa = 11.84TAPSDD169 pKa = 3.6VLQGEE174 pKa = 4.35VHH176 pKa = 6.59GNFLGEE182 pKa = 4.92LGHH185 pKa = 5.7QTLGLIVLNDD195 pKa = 3.6AYY197 pKa = 10.34GTGLAKK203 pKa = 10.77YY204 pKa = 8.47ITEE207 pKa = 4.17AFEE210 pKa = 4.32GAGGEE215 pKa = 4.5VVASEE220 pKa = 4.39TFNPGDD226 pKa = 3.6TSFDD230 pKa = 3.61SQIAAVLAEE239 pKa = 5.17DD240 pKa = 4.36PDD242 pKa = 5.98AITVISFDD250 pKa = 3.56EE251 pKa = 4.73AKK253 pKa = 10.12TILPSLIEE261 pKa = 4.12KK262 pKa = 10.78GFDD265 pKa = 3.25AEE267 pKa = 4.85NLFLVDD273 pKa = 4.63GNMAAYY279 pKa = 10.28DD280 pKa = 3.75KK281 pKa = 10.96DD282 pKa = 4.19FPAGLLEE289 pKa = 4.51GAKK292 pKa = 8.77GTYY295 pKa = 9.39PGPTPDD301 pKa = 3.18QVQEE305 pKa = 4.19FVDD308 pKa = 3.84GLNAFVEE315 pKa = 4.64EE316 pKa = 4.76SGTEE320 pKa = 3.8ALSDD324 pKa = 3.39YY325 pKa = 10.26TYY327 pKa = 11.26APEE330 pKa = 5.11SFDD333 pKa = 3.71AVNLIALASLAAQSTEE349 pKa = 3.83PADD352 pKa = 3.39IAEE355 pKa = 4.49KK356 pKa = 10.35LVEE359 pKa = 4.02VSGGSGEE366 pKa = 4.46GEE368 pKa = 4.2KK369 pKa = 9.9CTSFADD375 pKa = 3.8CADD378 pKa = 4.17IILGGGVADD387 pKa = 3.97YY388 pKa = 10.9DD389 pKa = 4.25GASSPVTFDD398 pKa = 3.31EE399 pKa = 4.97VGDD402 pKa = 3.89PTEE405 pKa = 4.02GMINIYY411 pKa = 10.38EE412 pKa = 4.03YY413 pKa = 11.22GADD416 pKa = 3.43NKK418 pKa = 10.19YY419 pKa = 10.76VPYY422 pKa = 10.56EE423 pKa = 3.99GG424 pKa = 4.4
Molecular weight: 43.73 kDa
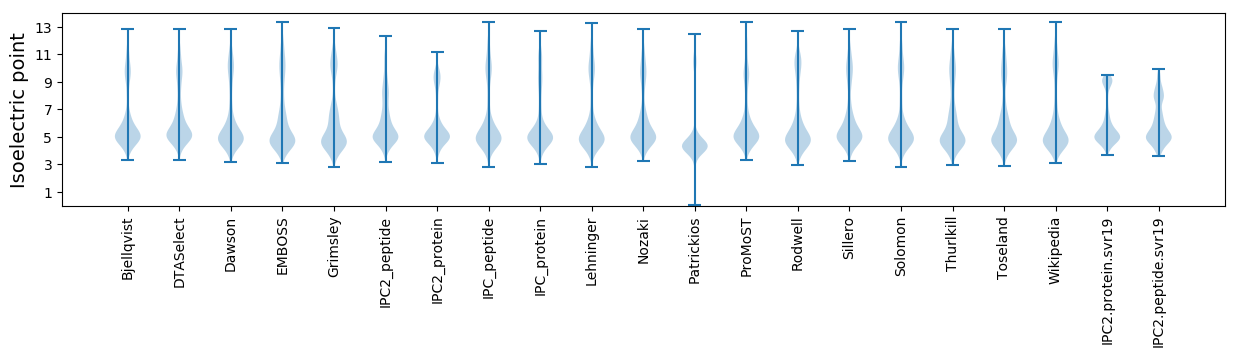
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9FU96|A0A4Y9FU96_9MICO YhgE/Pip domain-containing protein OS=Microbacterium paludicola OX=300019 GN=E4U02_08900 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
972793 |
32 |
2067 |
323.6 |
34.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.693 ± 0.061 | 0.489 ± 0.01 |
6.322 ± 0.035 | 6.129 ± 0.042 |
3.038 ± 0.029 | 8.938 ± 0.041 |
2.084 ± 0.022 | 4.567 ± 0.032 |
1.966 ± 0.034 | 10.038 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.777 ± 0.018 | 1.813 ± 0.024 |
5.517 ± 0.032 | 2.772 ± 0.023 |
7.723 ± 0.061 | 5.122 ± 0.029 |
5.828 ± 0.036 | 8.67 ± 0.045 |
1.536 ± 0.019 | 1.975 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
