
Wheat yellow striate virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Alphanucleorhabdovirus; Wheat yellow striate alphanucleorhabdovirus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
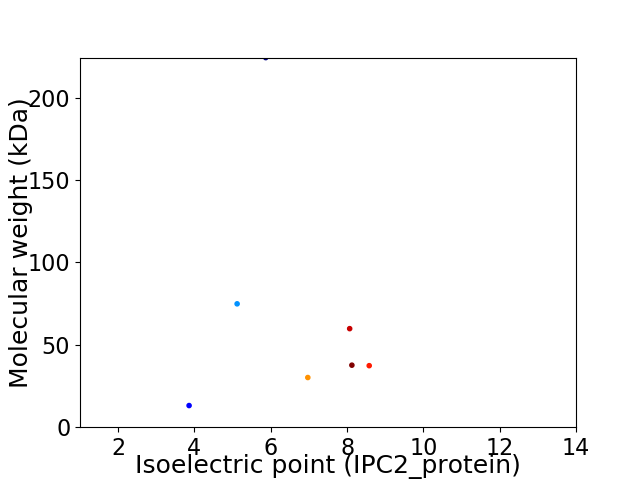
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
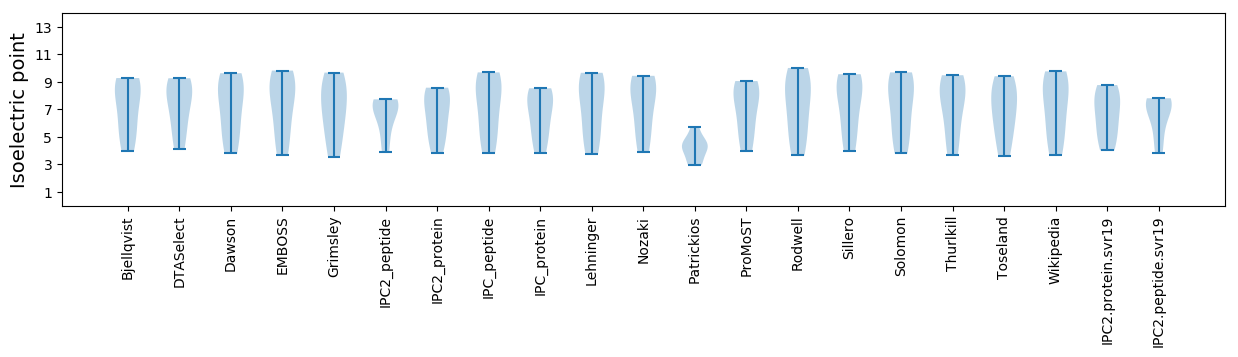
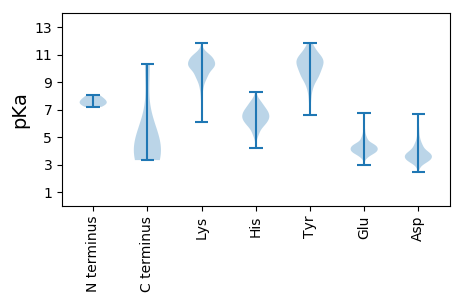
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R4K2H4|A0A2R4K2H4_9RHAB Nucleocapsid protein OS=Wheat yellow striate virus OX=2152660 PE=4 SV=1
MM1 pKa = 8.05DD2 pKa = 5.28KK3 pKa = 11.07SLDD6 pKa = 3.82NKK8 pKa = 10.62QGDD11 pKa = 4.45LLKK14 pKa = 10.07MSKK17 pKa = 8.38PTADD21 pKa = 3.3TGSPDD26 pKa = 3.65TPHH29 pKa = 7.66PEE31 pKa = 3.98PAHH34 pKa = 5.51TPEE37 pKa = 5.57GDD39 pKa = 3.29TTQKK43 pKa = 11.02EE44 pKa = 4.32DD45 pKa = 3.36TKK47 pKa = 11.51GNTGKK52 pKa = 10.64EE53 pKa = 3.87EE54 pKa = 3.96AAQSPEE60 pKa = 4.03AEE62 pKa = 4.48QEE64 pKa = 4.39DD65 pKa = 4.14DD66 pKa = 4.7TQPEE70 pKa = 4.37WCCEE74 pKa = 3.71IEE76 pKa = 4.13EE77 pKa = 5.12LDD79 pKa = 5.01VEE81 pKa = 4.66SDD83 pKa = 3.1WEE85 pKa = 4.03FCYY88 pKa = 10.58RR89 pKa = 11.84EE90 pKa = 4.19EE91 pKa = 6.18DD92 pKa = 3.47FDD94 pKa = 5.99CYY96 pKa = 10.81FDD98 pKa = 4.18NVWVYY103 pKa = 10.73EE104 pKa = 5.27LIDD107 pKa = 3.61EE108 pKa = 4.56CNGWRR113 pKa = 11.84EE114 pKa = 3.82
MM1 pKa = 8.05DD2 pKa = 5.28KK3 pKa = 11.07SLDD6 pKa = 3.82NKK8 pKa = 10.62QGDD11 pKa = 4.45LLKK14 pKa = 10.07MSKK17 pKa = 8.38PTADD21 pKa = 3.3TGSPDD26 pKa = 3.65TPHH29 pKa = 7.66PEE31 pKa = 3.98PAHH34 pKa = 5.51TPEE37 pKa = 5.57GDD39 pKa = 3.29TTQKK43 pKa = 11.02EE44 pKa = 4.32DD45 pKa = 3.36TKK47 pKa = 11.51GNTGKK52 pKa = 10.64EE53 pKa = 3.87EE54 pKa = 3.96AAQSPEE60 pKa = 4.03AEE62 pKa = 4.48QEE64 pKa = 4.39DD65 pKa = 4.14DD66 pKa = 4.7TQPEE70 pKa = 4.37WCCEE74 pKa = 3.71IEE76 pKa = 4.13EE77 pKa = 5.12LDD79 pKa = 5.01VEE81 pKa = 4.66SDD83 pKa = 3.1WEE85 pKa = 4.03FCYY88 pKa = 10.58RR89 pKa = 11.84EE90 pKa = 4.19EE91 pKa = 6.18DD92 pKa = 3.47FDD94 pKa = 5.99CYY96 pKa = 10.81FDD98 pKa = 4.18NVWVYY103 pKa = 10.73EE104 pKa = 5.27LIDD107 pKa = 3.61EE108 pKa = 4.56CNGWRR113 pKa = 11.84EE114 pKa = 3.82
Molecular weight: 13.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R4K2I6|A0A2R4K2I6_9RHAB GDP polyribonucleotidyltransferase OS=Wheat yellow striate virus OX=2152660 PE=4 SV=1
MM1 pKa = 7.85ADD3 pKa = 3.76SLDD6 pKa = 3.69TDD8 pKa = 3.84RR9 pKa = 11.84GRR11 pKa = 11.84TTRR14 pKa = 11.84SGSKK18 pKa = 10.26LKK20 pKa = 10.75AQTTHH25 pKa = 6.99RR26 pKa = 11.84VGTSSGSVRR35 pKa = 11.84GGKK38 pKa = 9.44PYY40 pKa = 10.83EE41 pKa = 4.15KK42 pKa = 10.31DD43 pKa = 3.56FKK45 pKa = 10.91AVEE48 pKa = 4.02ARR50 pKa = 11.84FHH52 pKa = 6.28NFDD55 pKa = 5.6PISASIIRR63 pKa = 11.84GDD65 pKa = 4.02QEE67 pKa = 3.97GTKK70 pKa = 10.41ADD72 pKa = 5.09LIMSDD77 pKa = 3.44SSVTKK82 pKa = 9.56ATAPAEE88 pKa = 4.24AAAPPPPPQPAAVPLTPPASATTQGQKK115 pKa = 10.36RR116 pKa = 11.84PGDD119 pKa = 3.76PEE121 pKa = 3.94TLEE124 pKa = 4.23EE125 pKa = 4.23GNAAKK130 pKa = 10.2RR131 pKa = 11.84VQRR134 pKa = 11.84ANKK137 pKa = 9.22VNNLLTTQGIGEE149 pKa = 4.22PSKK152 pKa = 11.01SAISQYY158 pKa = 10.69VVARR162 pKa = 11.84LNANNVEE169 pKa = 4.36HH170 pKa = 7.67DD171 pKa = 3.81DD172 pKa = 4.58AMVAEE177 pKa = 4.78CANMAVYY184 pKa = 9.67GWKK187 pKa = 9.88EE188 pKa = 3.61GKK190 pKa = 10.3KK191 pKa = 9.94FVSTQILNQATTVIPDD207 pKa = 5.42LITSMVTNANLITNAANALNSIPDD231 pKa = 3.69KK232 pKa = 10.65VAGAIRR238 pKa = 11.84TNIEE242 pKa = 3.47HH243 pKa = 6.79VAFQTGSKK251 pKa = 8.26ATKK254 pKa = 9.48RR255 pKa = 11.84DD256 pKa = 3.52TLVRR260 pKa = 11.84TAEE263 pKa = 4.28SIYY266 pKa = 10.75QNAAAEE272 pKa = 4.41SKK274 pKa = 11.17VDD276 pKa = 4.46FINNFIISAGINIQEE291 pKa = 4.54VKK293 pKa = 10.74RR294 pKa = 11.84NVANYY299 pKa = 7.88RR300 pKa = 11.84TIANAIIKK308 pKa = 10.26RR309 pKa = 11.84NTVLNIINEE318 pKa = 4.49TTEE321 pKa = 3.93HH322 pKa = 6.04QALLTQVQGNKK333 pKa = 10.16DD334 pKa = 4.29DD335 pKa = 3.43IRR337 pKa = 11.84NIARR341 pKa = 11.84GLSASYY347 pKa = 10.57VII349 pKa = 5.25
MM1 pKa = 7.85ADD3 pKa = 3.76SLDD6 pKa = 3.69TDD8 pKa = 3.84RR9 pKa = 11.84GRR11 pKa = 11.84TTRR14 pKa = 11.84SGSKK18 pKa = 10.26LKK20 pKa = 10.75AQTTHH25 pKa = 6.99RR26 pKa = 11.84VGTSSGSVRR35 pKa = 11.84GGKK38 pKa = 9.44PYY40 pKa = 10.83EE41 pKa = 4.15KK42 pKa = 10.31DD43 pKa = 3.56FKK45 pKa = 10.91AVEE48 pKa = 4.02ARR50 pKa = 11.84FHH52 pKa = 6.28NFDD55 pKa = 5.6PISASIIRR63 pKa = 11.84GDD65 pKa = 4.02QEE67 pKa = 3.97GTKK70 pKa = 10.41ADD72 pKa = 5.09LIMSDD77 pKa = 3.44SSVTKK82 pKa = 9.56ATAPAEE88 pKa = 4.24AAAPPPPPQPAAVPLTPPASATTQGQKK115 pKa = 10.36RR116 pKa = 11.84PGDD119 pKa = 3.76PEE121 pKa = 3.94TLEE124 pKa = 4.23EE125 pKa = 4.23GNAAKK130 pKa = 10.2RR131 pKa = 11.84VQRR134 pKa = 11.84ANKK137 pKa = 9.22VNNLLTTQGIGEE149 pKa = 4.22PSKK152 pKa = 11.01SAISQYY158 pKa = 10.69VVARR162 pKa = 11.84LNANNVEE169 pKa = 4.36HH170 pKa = 7.67DD171 pKa = 3.81DD172 pKa = 4.58AMVAEE177 pKa = 4.78CANMAVYY184 pKa = 9.67GWKK187 pKa = 9.88EE188 pKa = 3.61GKK190 pKa = 10.3KK191 pKa = 9.94FVSTQILNQATTVIPDD207 pKa = 5.42LITSMVTNANLITNAANALNSIPDD231 pKa = 3.69KK232 pKa = 10.65VAGAIRR238 pKa = 11.84TNIEE242 pKa = 3.47HH243 pKa = 6.79VAFQTGSKK251 pKa = 8.26ATKK254 pKa = 9.48RR255 pKa = 11.84DD256 pKa = 3.52TLVRR260 pKa = 11.84TAEE263 pKa = 4.28SIYY266 pKa = 10.75QNAAAEE272 pKa = 4.41SKK274 pKa = 11.17VDD276 pKa = 4.46FINNFIISAGINIQEE291 pKa = 4.54VKK293 pKa = 10.74RR294 pKa = 11.84NVANYY299 pKa = 7.88RR300 pKa = 11.84TIANAIIKK308 pKa = 10.26RR309 pKa = 11.84NTVLNIINEE318 pKa = 4.49TTEE321 pKa = 3.93HH322 pKa = 6.04QALLTQVQGNKK333 pKa = 10.16DD334 pKa = 4.29DD335 pKa = 3.43IRR337 pKa = 11.84NIARR341 pKa = 11.84GLSASYY347 pKa = 10.57VII349 pKa = 5.25
Molecular weight: 37.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4238 |
114 |
1965 |
605.4 |
68.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.521 ± 1.353 | 1.628 ± 0.296 |
6.111 ± 0.509 | 5.757 ± 0.681 |
3.115 ± 0.246 | 6.371 ± 0.351 |
2.076 ± 0.236 | 7.315 ± 0.64 |
5.781 ± 0.462 | 8.66 ± 0.843 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.539 ± 0.345 | 5.002 ± 0.48 |
4.625 ± 0.643 | 3.728 ± 0.673 |
5.05 ± 0.439 | 7.551 ± 0.88 |
6.914 ± 0.497 | 5.923 ± 0.204 |
1.534 ± 0.228 | 3.799 ± 0.353 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
