
Geodermatophilus sp. LHW52908
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus; unclassified Geodermatophilus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
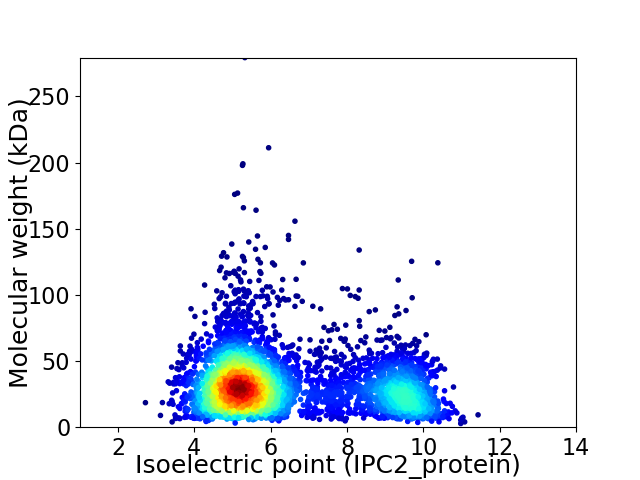
Virtual 2D-PAGE plot for 4784 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A372IY23|A0A372IY23_9ACTN Uncharacterized protein OS=Geodermatophilus sp. LHW52908 OX=2303986 GN=D0Z06_18850 PE=4 SV=1
MM1 pKa = 7.37RR2 pKa = 11.84HH3 pKa = 4.91VLSRR7 pKa = 11.84AIAAAAVGSVLLAGCSSTVTGTASAARR34 pKa = 11.84PVTDD38 pKa = 3.3VTAEE42 pKa = 3.95EE43 pKa = 4.34FLVTGATDD51 pKa = 3.57SDD53 pKa = 3.08IDD55 pKa = 3.75RR56 pKa = 11.84YY57 pKa = 10.77ARR59 pKa = 11.84NALADD64 pKa = 3.64LTTFWEE70 pKa = 3.95QAYY73 pKa = 9.68PEE75 pKa = 4.87FYY77 pKa = 10.99GEE79 pKa = 4.84PLTPLQGGYY88 pKa = 10.49FSVDD92 pKa = 2.74ADD94 pKa = 4.93DD95 pKa = 4.66IDD97 pKa = 3.8EE98 pKa = 4.29SRR100 pKa = 11.84YY101 pKa = 9.21PPTGVGCARR110 pKa = 11.84LPIDD114 pKa = 3.62PAEE117 pKa = 4.19VEE119 pKa = 4.39GNAFYY124 pKa = 10.91EE125 pKa = 4.44LNCDD129 pKa = 3.79VIVYY133 pKa = 9.96DD134 pKa = 4.69SSLLQEE140 pKa = 4.68LGEE143 pKa = 3.94LHH145 pKa = 7.03GRR147 pKa = 11.84FLGPAVMAHH156 pKa = 6.07EE157 pKa = 5.65FGHH160 pKa = 6.59AMQGRR165 pKa = 11.84FGFSEE170 pKa = 3.89VTIRR174 pKa = 11.84DD175 pKa = 3.63EE176 pKa = 4.19TQADD180 pKa = 4.0CFAGAFTRR188 pKa = 11.84WVVDD192 pKa = 3.6GNAEE196 pKa = 4.07HH197 pKa = 6.23VALRR201 pKa = 11.84VSEE204 pKa = 4.5LDD206 pKa = 3.56DD207 pKa = 3.49VLLGFIEE214 pKa = 4.44LRR216 pKa = 11.84DD217 pKa = 3.81PVGLTDD223 pKa = 3.54EE224 pKa = 4.59TVEE227 pKa = 4.27GAHH230 pKa = 6.45GSGFDD235 pKa = 3.47RR236 pKa = 11.84VSGFYY241 pKa = 10.3AGYY244 pKa = 7.7TGGVGTCRR252 pKa = 11.84DD253 pKa = 3.38EE254 pKa = 4.58FGPEE258 pKa = 3.61RR259 pKa = 11.84VFTAAEE265 pKa = 3.61FDD267 pKa = 3.77EE268 pKa = 5.99AEE270 pKa = 4.27EE271 pKa = 4.06ATEE274 pKa = 4.36GNADD278 pKa = 3.5YY279 pKa = 11.62ADD281 pKa = 3.26IQEE284 pKa = 4.27IVEE287 pKa = 4.13LSLPAFYY294 pKa = 10.61DD295 pKa = 3.1SWFPQVGGAAFDD307 pKa = 3.87APAIEE312 pKa = 4.97PFDD315 pKa = 3.89GTAPACGDD323 pKa = 3.36MGAEE327 pKa = 4.0DD328 pKa = 4.67RR329 pKa = 11.84DD330 pKa = 4.07LGFCAADD337 pKa = 3.29STVYY341 pKa = 10.97ADD343 pKa = 3.71EE344 pKa = 4.61TDD346 pKa = 3.81LLRR349 pKa = 11.84PAYY352 pKa = 10.25QEE354 pKa = 4.17FGDD357 pKa = 4.22FAVATAISLPYY368 pKa = 9.77AQAARR373 pKa = 11.84AQLGLSTDD381 pKa = 3.89DD382 pKa = 3.48SAGTISTVCLTGWYY396 pKa = 9.59AARR399 pKa = 11.84FFDD402 pKa = 5.5GDD404 pKa = 3.59FADD407 pKa = 4.55TEE409 pKa = 4.38LSPGDD414 pKa = 3.47VDD416 pKa = 4.06EE417 pKa = 6.64AILFLLGHH425 pKa = 6.05GQDD428 pKa = 4.95DD429 pKa = 4.02SVLPNVEE436 pKa = 3.89LSGFEE441 pKa = 3.75LVGAFRR447 pKa = 11.84DD448 pKa = 4.01GFLQGGTACDD458 pKa = 3.73LGVV461 pKa = 3.23
MM1 pKa = 7.37RR2 pKa = 11.84HH3 pKa = 4.91VLSRR7 pKa = 11.84AIAAAAVGSVLLAGCSSTVTGTASAARR34 pKa = 11.84PVTDD38 pKa = 3.3VTAEE42 pKa = 3.95EE43 pKa = 4.34FLVTGATDD51 pKa = 3.57SDD53 pKa = 3.08IDD55 pKa = 3.75RR56 pKa = 11.84YY57 pKa = 10.77ARR59 pKa = 11.84NALADD64 pKa = 3.64LTTFWEE70 pKa = 3.95QAYY73 pKa = 9.68PEE75 pKa = 4.87FYY77 pKa = 10.99GEE79 pKa = 4.84PLTPLQGGYY88 pKa = 10.49FSVDD92 pKa = 2.74ADD94 pKa = 4.93DD95 pKa = 4.66IDD97 pKa = 3.8EE98 pKa = 4.29SRR100 pKa = 11.84YY101 pKa = 9.21PPTGVGCARR110 pKa = 11.84LPIDD114 pKa = 3.62PAEE117 pKa = 4.19VEE119 pKa = 4.39GNAFYY124 pKa = 10.91EE125 pKa = 4.44LNCDD129 pKa = 3.79VIVYY133 pKa = 9.96DD134 pKa = 4.69SSLLQEE140 pKa = 4.68LGEE143 pKa = 3.94LHH145 pKa = 7.03GRR147 pKa = 11.84FLGPAVMAHH156 pKa = 6.07EE157 pKa = 5.65FGHH160 pKa = 6.59AMQGRR165 pKa = 11.84FGFSEE170 pKa = 3.89VTIRR174 pKa = 11.84DD175 pKa = 3.63EE176 pKa = 4.19TQADD180 pKa = 4.0CFAGAFTRR188 pKa = 11.84WVVDD192 pKa = 3.6GNAEE196 pKa = 4.07HH197 pKa = 6.23VALRR201 pKa = 11.84VSEE204 pKa = 4.5LDD206 pKa = 3.56DD207 pKa = 3.49VLLGFIEE214 pKa = 4.44LRR216 pKa = 11.84DD217 pKa = 3.81PVGLTDD223 pKa = 3.54EE224 pKa = 4.59TVEE227 pKa = 4.27GAHH230 pKa = 6.45GSGFDD235 pKa = 3.47RR236 pKa = 11.84VSGFYY241 pKa = 10.3AGYY244 pKa = 7.7TGGVGTCRR252 pKa = 11.84DD253 pKa = 3.38EE254 pKa = 4.58FGPEE258 pKa = 3.61RR259 pKa = 11.84VFTAAEE265 pKa = 3.61FDD267 pKa = 3.77EE268 pKa = 5.99AEE270 pKa = 4.27EE271 pKa = 4.06ATEE274 pKa = 4.36GNADD278 pKa = 3.5YY279 pKa = 11.62ADD281 pKa = 3.26IQEE284 pKa = 4.27IVEE287 pKa = 4.13LSLPAFYY294 pKa = 10.61DD295 pKa = 3.1SWFPQVGGAAFDD307 pKa = 3.87APAIEE312 pKa = 4.97PFDD315 pKa = 3.89GTAPACGDD323 pKa = 3.36MGAEE327 pKa = 4.0DD328 pKa = 4.67RR329 pKa = 11.84DD330 pKa = 4.07LGFCAADD337 pKa = 3.29STVYY341 pKa = 10.97ADD343 pKa = 3.71EE344 pKa = 4.61TDD346 pKa = 3.81LLRR349 pKa = 11.84PAYY352 pKa = 10.25QEE354 pKa = 4.17FGDD357 pKa = 4.22FAVATAISLPYY368 pKa = 9.77AQAARR373 pKa = 11.84AQLGLSTDD381 pKa = 3.89DD382 pKa = 3.48SAGTISTVCLTGWYY396 pKa = 9.59AARR399 pKa = 11.84FFDD402 pKa = 5.5GDD404 pKa = 3.59FADD407 pKa = 4.55TEE409 pKa = 4.38LSPGDD414 pKa = 3.47VDD416 pKa = 4.06EE417 pKa = 6.64AILFLLGHH425 pKa = 6.05GQDD428 pKa = 4.95DD429 pKa = 4.02SVLPNVEE436 pKa = 3.89LSGFEE441 pKa = 3.75LVGAFRR447 pKa = 11.84DD448 pKa = 4.01GFLQGGTACDD458 pKa = 3.73LGVV461 pKa = 3.23
Molecular weight: 49.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A372IXK3|A0A372IXK3_9ACTN Uncharacterized protein OS=Geodermatophilus sp. LHW52908 OX=2303986 GN=D0Z06_23070 PE=4 SV=1
MM1 pKa = 7.57PHH3 pKa = 7.03RR4 pKa = 11.84PQPASSRR11 pKa = 11.84TAAPRR16 pKa = 11.84TVPRR20 pKa = 11.84STARR24 pKa = 11.84PSTAPPSTAPPSTAPPSTAPRR45 pKa = 11.84STAHH49 pKa = 7.24RR50 pKa = 11.84STGPSPTAPRR60 pKa = 11.84RR61 pKa = 11.84TGPRR65 pKa = 11.84RR66 pKa = 11.84AGGTAPPVTPRR77 pKa = 11.84PRR79 pKa = 11.84RR80 pKa = 11.84TPAPSRR86 pKa = 11.84PRR88 pKa = 11.84ARR90 pKa = 3.61
MM1 pKa = 7.57PHH3 pKa = 7.03RR4 pKa = 11.84PQPASSRR11 pKa = 11.84TAAPRR16 pKa = 11.84TVPRR20 pKa = 11.84STARR24 pKa = 11.84PSTAPPSTAPPSTAPPSTAPRR45 pKa = 11.84STAHH49 pKa = 7.24RR50 pKa = 11.84STGPSPTAPRR60 pKa = 11.84RR61 pKa = 11.84TGPRR65 pKa = 11.84RR66 pKa = 11.84AGGTAPPVTPRR77 pKa = 11.84PRR79 pKa = 11.84RR80 pKa = 11.84TPAPSRR86 pKa = 11.84PRR88 pKa = 11.84ARR90 pKa = 3.61
Molecular weight: 9.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1526184 |
24 |
2587 |
319.0 |
33.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.01 ± 0.054 | 0.698 ± 0.009 |
6.082 ± 0.027 | 5.535 ± 0.034 |
2.474 ± 0.022 | 10.026 ± 0.033 |
2.063 ± 0.016 | 2.346 ± 0.025 |
1.113 ± 0.019 | 10.761 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.486 ± 0.013 | 1.261 ± 0.016 |
6.562 ± 0.04 | 2.463 ± 0.019 |
8.817 ± 0.039 | 4.333 ± 0.021 |
5.766 ± 0.028 | 10.0 ± 0.04 |
1.512 ± 0.018 | 1.691 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
