
Reptilian orthoreovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Orthoreovirus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
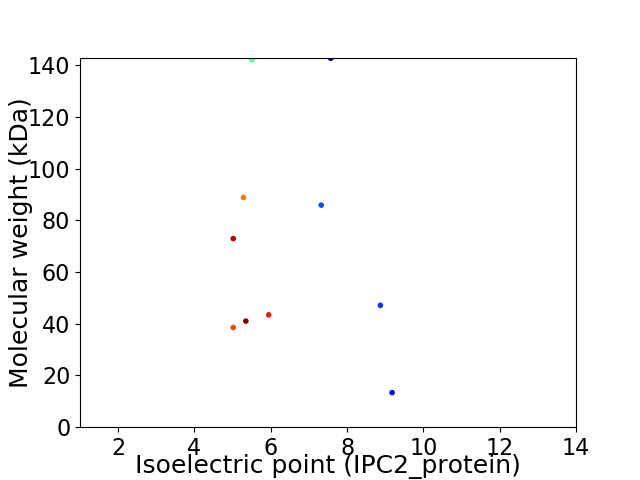
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|W8PTM7|W8PTM7_9REOV Sigma C OS=Reptilian orthoreovirus OX=226613 GN=SC PE=4 SV=1
MM1 pKa = 7.5GNASSLVQNFNITGDD16 pKa = 3.89GNSFAPTADD25 pKa = 3.52QTSSAVPTLSLQPGILNPGGLAFKK49 pKa = 10.79LIDD52 pKa = 3.7PTKK55 pKa = 10.91DD56 pKa = 3.06PTDD59 pKa = 4.24PGALAIMTTEE69 pKa = 4.73DD70 pKa = 4.02LPEE73 pKa = 4.07LAITDD78 pKa = 4.15TNNSAGALPISGMYY92 pKa = 10.2LDD94 pKa = 5.0SKK96 pKa = 10.33PEE98 pKa = 3.87KK99 pKa = 9.17LTVVTDD105 pKa = 3.72HH106 pKa = 7.31ALEE109 pKa = 3.88QMRR112 pKa = 11.84KK113 pKa = 8.86NEE115 pKa = 4.08SAVEE119 pKa = 3.7ISRR122 pKa = 11.84PYY124 pKa = 10.16LSRR127 pKa = 11.84IAVMPQSTFFTDD139 pKa = 3.44YY140 pKa = 9.91VVKK143 pKa = 10.75IKK145 pKa = 10.68GCYY148 pKa = 8.75VGASATQASRR158 pKa = 11.84TYY160 pKa = 10.79VEE162 pKa = 4.25GPYY165 pKa = 10.6VITQSRR171 pKa = 11.84MTTYY175 pKa = 10.09MSIMATASNALSKK188 pKa = 10.16WSRR191 pKa = 11.84AVTEE195 pKa = 4.09VLNIIPTTTPYY206 pKa = 11.37GKK208 pKa = 8.09TEE210 pKa = 3.99CDD212 pKa = 2.93MRR214 pKa = 11.84SIIKK218 pKa = 10.15FLDD221 pKa = 3.25QVLPEE226 pKa = 4.11DD227 pKa = 4.09HH228 pKa = 7.12LARR231 pKa = 11.84DD232 pKa = 4.07YY233 pKa = 10.99PEE235 pKa = 4.79EE236 pKa = 3.87MAEE239 pKa = 4.21TVAKK243 pKa = 10.39INGGIRR249 pKa = 11.84WKK251 pKa = 11.03DD252 pKa = 3.51EE253 pKa = 3.96LTGKK257 pKa = 8.67ATSLAINDD265 pKa = 3.43VAAPTMATLSNTMPLAEE282 pKa = 4.56KK283 pKa = 10.87SEE285 pKa = 4.31TTTDD289 pKa = 3.47SLSLINDD296 pKa = 3.57ADD298 pKa = 4.07VDD300 pKa = 4.38LMSCDD305 pKa = 3.74RR306 pKa = 11.84PISSTVWARR315 pKa = 11.84TVEE318 pKa = 4.06PKK320 pKa = 10.48DD321 pKa = 3.43YY322 pKa = 10.54NIRR325 pKa = 11.84VLPLEE330 pKa = 3.83ASMWLRR336 pKa = 11.84TAALTANFVVSWKK349 pKa = 10.42SKK351 pKa = 10.61EE352 pKa = 4.03SDD354 pKa = 2.69ATNYY358 pKa = 9.32IKK360 pKa = 10.55IMTGARR366 pKa = 11.84VINLEE371 pKa = 3.86QTGTMTFTVDD381 pKa = 5.31LEE383 pKa = 4.31GKK385 pKa = 9.58NYY387 pKa = 7.99KK388 pKa = 7.72TTSFDD393 pKa = 4.12PNGKK397 pKa = 7.48TVGLVAMQSKK407 pKa = 9.86VPFEE411 pKa = 3.98QWTAASMVIGATMVASEE428 pKa = 4.64VLVAAQSSTPGASIIAKK445 pKa = 8.47TALTYY450 pKa = 10.25IFEE453 pKa = 4.82RR454 pKa = 11.84EE455 pKa = 4.29TIPLADD461 pKa = 3.92TEE463 pKa = 4.66VNTYY467 pKa = 9.03LFCGFVLTEE476 pKa = 4.36TPTEE480 pKa = 4.43SNYY483 pKa = 10.06PWYY486 pKa = 9.97DD487 pKa = 2.8VWDD490 pKa = 3.89ATTTLTPMTTGTVTVNGSTVNEE512 pKa = 4.16VVPTSLIGAYY522 pKa = 8.91TPTAMSAALPNDD534 pKa = 3.49AGVILQGRR542 pKa = 11.84AEE544 pKa = 4.33KK545 pKa = 10.23LAKK548 pKa = 10.22AIKK551 pKa = 10.71NEE553 pKa = 4.14DD554 pKa = 3.57DD555 pKa = 3.79SVADD559 pKa = 3.79EE560 pKa = 4.45ASTYY564 pKa = 11.13SMPIMGVLALQDD576 pKa = 3.38AGKK579 pKa = 8.21PTMRR583 pKa = 11.84AKK585 pKa = 10.46FDD587 pKa = 3.81PVGLLKK593 pKa = 10.53KK594 pKa = 10.56ASRR597 pKa = 11.84AFQMFLGNPKK607 pKa = 10.31SILDD611 pKa = 3.43MTTPVMKK618 pKa = 10.71DD619 pKa = 3.12PAVWRR624 pKa = 11.84ALAQGVRR631 pKa = 11.84DD632 pKa = 4.77GIRR635 pKa = 11.84NKK637 pKa = 10.97SMGAGVKK644 pKa = 10.46SIFDD648 pKa = 3.6KK649 pKa = 11.37VSATKK654 pKa = 10.31SVQKK658 pKa = 9.71WKK660 pKa = 10.75QGILTQIQTLYY671 pKa = 9.69PPSGEE676 pKa = 3.99
MM1 pKa = 7.5GNASSLVQNFNITGDD16 pKa = 3.89GNSFAPTADD25 pKa = 3.52QTSSAVPTLSLQPGILNPGGLAFKK49 pKa = 10.79LIDD52 pKa = 3.7PTKK55 pKa = 10.91DD56 pKa = 3.06PTDD59 pKa = 4.24PGALAIMTTEE69 pKa = 4.73DD70 pKa = 4.02LPEE73 pKa = 4.07LAITDD78 pKa = 4.15TNNSAGALPISGMYY92 pKa = 10.2LDD94 pKa = 5.0SKK96 pKa = 10.33PEE98 pKa = 3.87KK99 pKa = 9.17LTVVTDD105 pKa = 3.72HH106 pKa = 7.31ALEE109 pKa = 3.88QMRR112 pKa = 11.84KK113 pKa = 8.86NEE115 pKa = 4.08SAVEE119 pKa = 3.7ISRR122 pKa = 11.84PYY124 pKa = 10.16LSRR127 pKa = 11.84IAVMPQSTFFTDD139 pKa = 3.44YY140 pKa = 9.91VVKK143 pKa = 10.75IKK145 pKa = 10.68GCYY148 pKa = 8.75VGASATQASRR158 pKa = 11.84TYY160 pKa = 10.79VEE162 pKa = 4.25GPYY165 pKa = 10.6VITQSRR171 pKa = 11.84MTTYY175 pKa = 10.09MSIMATASNALSKK188 pKa = 10.16WSRR191 pKa = 11.84AVTEE195 pKa = 4.09VLNIIPTTTPYY206 pKa = 11.37GKK208 pKa = 8.09TEE210 pKa = 3.99CDD212 pKa = 2.93MRR214 pKa = 11.84SIIKK218 pKa = 10.15FLDD221 pKa = 3.25QVLPEE226 pKa = 4.11DD227 pKa = 4.09HH228 pKa = 7.12LARR231 pKa = 11.84DD232 pKa = 4.07YY233 pKa = 10.99PEE235 pKa = 4.79EE236 pKa = 3.87MAEE239 pKa = 4.21TVAKK243 pKa = 10.39INGGIRR249 pKa = 11.84WKK251 pKa = 11.03DD252 pKa = 3.51EE253 pKa = 3.96LTGKK257 pKa = 8.67ATSLAINDD265 pKa = 3.43VAAPTMATLSNTMPLAEE282 pKa = 4.56KK283 pKa = 10.87SEE285 pKa = 4.31TTTDD289 pKa = 3.47SLSLINDD296 pKa = 3.57ADD298 pKa = 4.07VDD300 pKa = 4.38LMSCDD305 pKa = 3.74RR306 pKa = 11.84PISSTVWARR315 pKa = 11.84TVEE318 pKa = 4.06PKK320 pKa = 10.48DD321 pKa = 3.43YY322 pKa = 10.54NIRR325 pKa = 11.84VLPLEE330 pKa = 3.83ASMWLRR336 pKa = 11.84TAALTANFVVSWKK349 pKa = 10.42SKK351 pKa = 10.61EE352 pKa = 4.03SDD354 pKa = 2.69ATNYY358 pKa = 9.32IKK360 pKa = 10.55IMTGARR366 pKa = 11.84VINLEE371 pKa = 3.86QTGTMTFTVDD381 pKa = 5.31LEE383 pKa = 4.31GKK385 pKa = 9.58NYY387 pKa = 7.99KK388 pKa = 7.72TTSFDD393 pKa = 4.12PNGKK397 pKa = 7.48TVGLVAMQSKK407 pKa = 9.86VPFEE411 pKa = 3.98QWTAASMVIGATMVASEE428 pKa = 4.64VLVAAQSSTPGASIIAKK445 pKa = 8.47TALTYY450 pKa = 10.25IFEE453 pKa = 4.82RR454 pKa = 11.84EE455 pKa = 4.29TIPLADD461 pKa = 3.92TEE463 pKa = 4.66VNTYY467 pKa = 9.03LFCGFVLTEE476 pKa = 4.36TPTEE480 pKa = 4.43SNYY483 pKa = 10.06PWYY486 pKa = 9.97DD487 pKa = 2.8VWDD490 pKa = 3.89ATTTLTPMTTGTVTVNGSTVNEE512 pKa = 4.16VVPTSLIGAYY522 pKa = 8.91TPTAMSAALPNDD534 pKa = 3.49AGVILQGRR542 pKa = 11.84AEE544 pKa = 4.33KK545 pKa = 10.23LAKK548 pKa = 10.22AIKK551 pKa = 10.71NEE553 pKa = 4.14DD554 pKa = 3.57DD555 pKa = 3.79SVADD559 pKa = 3.79EE560 pKa = 4.45ASTYY564 pKa = 11.13SMPIMGVLALQDD576 pKa = 3.38AGKK579 pKa = 8.21PTMRR583 pKa = 11.84AKK585 pKa = 10.46FDD587 pKa = 3.81PVGLLKK593 pKa = 10.53KK594 pKa = 10.56ASRR597 pKa = 11.84AFQMFLGNPKK607 pKa = 10.31SILDD611 pKa = 3.43MTTPVMKK618 pKa = 10.71DD619 pKa = 3.12PAVWRR624 pKa = 11.84ALAQGVRR631 pKa = 11.84DD632 pKa = 4.77GIRR635 pKa = 11.84NKK637 pKa = 10.97SMGAGVKK644 pKa = 10.46SIFDD648 pKa = 3.6KK649 pKa = 11.37VSATKK654 pKa = 10.31SVQKK658 pKa = 9.71WKK660 pKa = 10.75QGILTQIQTLYY671 pKa = 9.69PPSGEE676 pKa = 3.99
Molecular weight: 72.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8P1A7|W8P1A7_9REOV Lambda C OS=Reptilian orthoreovirus OX=226613 GN=L2 PE=4 SV=1
MM1 pKa = 7.59GNGPSNFVNHH11 pKa = 6.27SPAEE15 pKa = 4.19TIISGLDD22 pKa = 2.97KK23 pKa = 11.26GADD26 pKa = 3.34KK27 pKa = 11.32VAGTVSNTIWEE38 pKa = 4.54VVGGVLLLLFLIAVGFGLYY57 pKa = 9.77RR58 pKa = 11.84YY59 pKa = 10.14AKK61 pKa = 9.73GRR63 pKa = 11.84RR64 pKa = 11.84EE65 pKa = 4.07KK66 pKa = 10.66KK67 pKa = 9.9RR68 pKa = 11.84EE69 pKa = 3.63LTEE72 pKa = 3.84FQKK75 pKa = 10.85RR76 pKa = 11.84FLRR79 pKa = 11.84NSYY82 pKa = 10.12RR83 pKa = 11.84LSQRR87 pKa = 11.84RR88 pKa = 11.84SLTQSPDD95 pKa = 3.32YY96 pKa = 10.85EE97 pKa = 4.4EE98 pKa = 4.15PTEE101 pKa = 4.01YY102 pKa = 10.82GITKK106 pKa = 9.05PLPPPPYY113 pKa = 8.93ATYY116 pKa = 10.94INII119 pKa = 4.03
MM1 pKa = 7.59GNGPSNFVNHH11 pKa = 6.27SPAEE15 pKa = 4.19TIISGLDD22 pKa = 2.97KK23 pKa = 11.26GADD26 pKa = 3.34KK27 pKa = 11.32VAGTVSNTIWEE38 pKa = 4.54VVGGVLLLLFLIAVGFGLYY57 pKa = 9.77RR58 pKa = 11.84YY59 pKa = 10.14AKK61 pKa = 9.73GRR63 pKa = 11.84RR64 pKa = 11.84EE65 pKa = 4.07KK66 pKa = 10.66KK67 pKa = 9.9RR68 pKa = 11.84EE69 pKa = 3.63LTEE72 pKa = 3.84FQKK75 pKa = 10.85RR76 pKa = 11.84FLRR79 pKa = 11.84NSYY82 pKa = 10.12RR83 pKa = 11.84LSQRR87 pKa = 11.84RR88 pKa = 11.84SLTQSPDD95 pKa = 3.32YY96 pKa = 10.85EE97 pKa = 4.4EE98 pKa = 4.15PTEE101 pKa = 4.01YY102 pKa = 10.82GITKK106 pKa = 9.05PLPPPPYY113 pKa = 8.93ATYY116 pKa = 10.94INII119 pKa = 4.03
Molecular weight: 13.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7699 |
119 |
1290 |
699.9 |
78.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.131 ± 0.616 | 1.221 ± 0.142 |
6.092 ± 0.321 | 4.429 ± 0.448 |
3.286 ± 0.172 | 5.221 ± 0.383 |
1.766 ± 0.284 | 5.663 ± 0.269 |
4.052 ± 0.548 | 9.469 ± 0.272 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.312 ± 0.272 | 4.546 ± 0.42 |
5.624 ± 0.346 | 4.234 ± 0.241 |
5.611 ± 0.312 | 7.715 ± 0.231 |
7.274 ± 0.486 | 7.17 ± 0.408 |
1.611 ± 0.127 | 3.572 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
