
Paraburkholderia humisilvae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
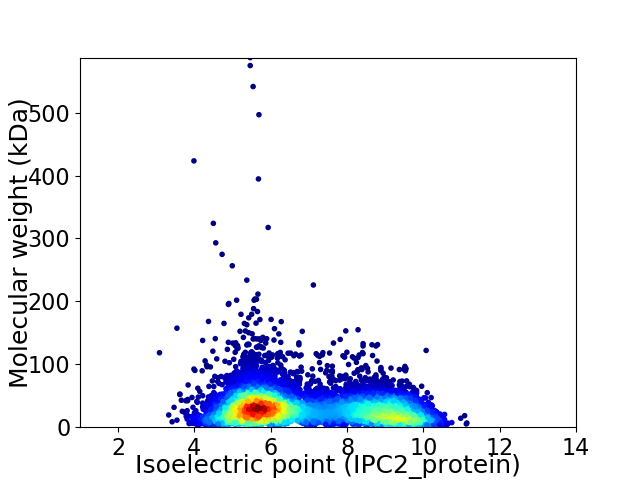
Virtual 2D-PAGE plot for 8698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J5EFD3|A0A6J5EFD3_9BURK Peripla_BP_6 domain-containing protein OS=Paraburkholderia humisilvae OX=627669 GN=LMG29542_05095 PE=4 SV=1
MM1 pKa = 6.89STVDD5 pKa = 3.53VFPSFNTDD13 pKa = 3.01MLSTPVDD20 pKa = 3.73SGSDD24 pKa = 3.67SPNLQQLEE32 pKa = 4.15QQYY35 pKa = 8.02MQLAQQFIQQGAGTLKK51 pKa = 8.85TQAAAGDD58 pKa = 3.85LASYY62 pKa = 7.53MTQDD66 pKa = 4.35NIGTLDD72 pKa = 3.76PNQLYY77 pKa = 10.79ALANNPPSGTPPGVSAAAAYY97 pKa = 7.73MLKK100 pKa = 10.44NSSAYY105 pKa = 9.27EE106 pKa = 3.81QIEE109 pKa = 4.26THH111 pKa = 6.87DD112 pKa = 3.68SSGADD117 pKa = 3.23GLSGVGNFQWASEE130 pKa = 4.31GGLSSASPFQMGSGSGFGGMSPFGGMPSMGGITAFGGMPSMGGMTSFGGMPSMGGMTSFGGMSSMGGMSSLGDD203 pKa = 3.32MSSFGDD209 pKa = 3.51MSSLFGSDD217 pKa = 3.23PLEE220 pKa = 4.26AQAAAGALASYY231 pKa = 9.47MSQNNIGALDD241 pKa = 3.99PNALYY246 pKa = 10.42QLANNPPPNMPPGVSEE262 pKa = 3.93AANYY266 pKa = 8.85MLEE269 pKa = 4.17NPDD272 pKa = 3.19IYY274 pKa = 10.98EE275 pKa = 4.02QIEE278 pKa = 4.19THH280 pKa = 6.68DD281 pKa = 3.55VAGADD286 pKa = 4.19GISGIGNFQWAAEE299 pKa = 4.38GGLMAGPAVSGSGGLPADD317 pKa = 4.49GGSGTDD323 pKa = 4.5DD324 pKa = 3.8PAEE327 pKa = 4.27NAPPANPSSSSGSTSGSSGGGTSSSTGGSTSGSKK361 pKa = 10.91GSDD364 pKa = 2.96STVPNISTSDD374 pKa = 3.58PAKK377 pKa = 10.71AIAEE381 pKa = 4.2DD382 pKa = 3.63LEE384 pKa = 4.14QRR386 pKa = 11.84YY387 pKa = 9.25GLTATQAAGVLGNLQQEE404 pKa = 4.65SGLQGDD410 pKa = 4.43VNQGGATGAPSSNDD424 pKa = 3.11ADD426 pKa = 4.53DD427 pKa = 5.25NGHH430 pKa = 5.9GWGLAQWGGTRR441 pKa = 11.84KK442 pKa = 9.83QGEE445 pKa = 3.6IDD447 pKa = 3.55YY448 pKa = 10.74ANEE451 pKa = 4.11HH452 pKa = 6.21GLDD455 pKa = 3.93PGSLQANIGFMNQEE469 pKa = 3.53LDD471 pKa = 3.31GAYY474 pKa = 10.46SKK476 pKa = 10.02TITDD480 pKa = 3.08IKK482 pKa = 9.51KK483 pKa = 8.43TNNVSDD489 pKa = 3.89AAKK492 pKa = 10.46VWDD495 pKa = 4.14EE496 pKa = 4.6DD497 pKa = 4.09YY498 pKa = 11.77EE499 pKa = 4.34EE500 pKa = 4.48ATDD503 pKa = 3.73PQMGNRR509 pKa = 11.84VQYY512 pKa = 10.23AQNFLDD518 pKa = 3.92EE519 pKa = 4.48GLL521 pKa = 3.94
MM1 pKa = 6.89STVDD5 pKa = 3.53VFPSFNTDD13 pKa = 3.01MLSTPVDD20 pKa = 3.73SGSDD24 pKa = 3.67SPNLQQLEE32 pKa = 4.15QQYY35 pKa = 8.02MQLAQQFIQQGAGTLKK51 pKa = 8.85TQAAAGDD58 pKa = 3.85LASYY62 pKa = 7.53MTQDD66 pKa = 4.35NIGTLDD72 pKa = 3.76PNQLYY77 pKa = 10.79ALANNPPSGTPPGVSAAAAYY97 pKa = 7.73MLKK100 pKa = 10.44NSSAYY105 pKa = 9.27EE106 pKa = 3.81QIEE109 pKa = 4.26THH111 pKa = 6.87DD112 pKa = 3.68SSGADD117 pKa = 3.23GLSGVGNFQWASEE130 pKa = 4.31GGLSSASPFQMGSGSGFGGMSPFGGMPSMGGITAFGGMPSMGGMTSFGGMPSMGGMTSFGGMSSMGGMSSLGDD203 pKa = 3.32MSSFGDD209 pKa = 3.51MSSLFGSDD217 pKa = 3.23PLEE220 pKa = 4.26AQAAAGALASYY231 pKa = 9.47MSQNNIGALDD241 pKa = 3.99PNALYY246 pKa = 10.42QLANNPPPNMPPGVSEE262 pKa = 3.93AANYY266 pKa = 8.85MLEE269 pKa = 4.17NPDD272 pKa = 3.19IYY274 pKa = 10.98EE275 pKa = 4.02QIEE278 pKa = 4.19THH280 pKa = 6.68DD281 pKa = 3.55VAGADD286 pKa = 4.19GISGIGNFQWAAEE299 pKa = 4.38GGLMAGPAVSGSGGLPADD317 pKa = 4.49GGSGTDD323 pKa = 4.5DD324 pKa = 3.8PAEE327 pKa = 4.27NAPPANPSSSSGSTSGSSGGGTSSSTGGSTSGSKK361 pKa = 10.91GSDD364 pKa = 2.96STVPNISTSDD374 pKa = 3.58PAKK377 pKa = 10.71AIAEE381 pKa = 4.2DD382 pKa = 3.63LEE384 pKa = 4.14QRR386 pKa = 11.84YY387 pKa = 9.25GLTATQAAGVLGNLQQEE404 pKa = 4.65SGLQGDD410 pKa = 4.43VNQGGATGAPSSNDD424 pKa = 3.11ADD426 pKa = 4.53DD427 pKa = 5.25NGHH430 pKa = 5.9GWGLAQWGGTRR441 pKa = 11.84KK442 pKa = 9.83QGEE445 pKa = 3.6IDD447 pKa = 3.55YY448 pKa = 10.74ANEE451 pKa = 4.11HH452 pKa = 6.21GLDD455 pKa = 3.93PGSLQANIGFMNQEE469 pKa = 3.53LDD471 pKa = 3.31GAYY474 pKa = 10.46SKK476 pKa = 10.02TITDD480 pKa = 3.08IKK482 pKa = 9.51KK483 pKa = 8.43TNNVSDD489 pKa = 3.89AAKK492 pKa = 10.46VWDD495 pKa = 4.14EE496 pKa = 4.6DD497 pKa = 4.09YY498 pKa = 11.77EE499 pKa = 4.34EE500 pKa = 4.48ATDD503 pKa = 3.73PQMGNRR509 pKa = 11.84VQYY512 pKa = 10.23AQNFLDD518 pKa = 3.92EE519 pKa = 4.48GLL521 pKa = 3.94
Molecular weight: 52.62 kDa
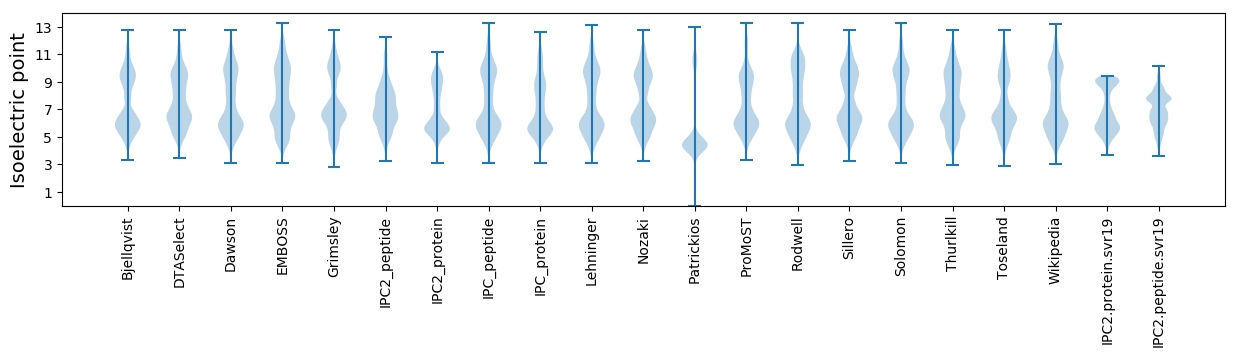
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J5DIH3|A0A6J5DIH3_9BURK STAS domain-containing protein OS=Paraburkholderia humisilvae OX=627669 GN=LMG29542_01941 PE=4 SV=1
MM1 pKa = 7.11HH2 pKa = 7.28TLLSTRR8 pKa = 11.84LTRR11 pKa = 11.84AALTAARR18 pKa = 11.84PARR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84VVRR26 pKa = 11.84ALRR29 pKa = 11.84HH30 pKa = 4.85HH31 pKa = 5.94SARR34 pKa = 11.84TRR36 pKa = 11.84ALTEE40 pKa = 3.48QWRR43 pKa = 11.84DD44 pKa = 3.16ARR46 pKa = 11.84PVDD49 pKa = 4.84SVRR52 pKa = 11.84TWFRR56 pKa = 11.84TFFSTLRR63 pKa = 11.84MQGASRR69 pKa = 11.84RR70 pKa = 11.84FSLRR74 pKa = 11.84TLLLRR79 pKa = 11.84RR80 pKa = 11.84PTPLRR85 pKa = 11.84VAGMAGGLNLVRR97 pKa = 11.84GLNVARR103 pKa = 11.84GMSRR107 pKa = 11.84KK108 pKa = 9.01PRR110 pKa = 11.84RR111 pKa = 11.84TSTTSTGWFAFARR124 pKa = 4.07
MM1 pKa = 7.11HH2 pKa = 7.28TLLSTRR8 pKa = 11.84LTRR11 pKa = 11.84AALTAARR18 pKa = 11.84PARR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84VVRR26 pKa = 11.84ALRR29 pKa = 11.84HH30 pKa = 4.85HH31 pKa = 5.94SARR34 pKa = 11.84TRR36 pKa = 11.84ALTEE40 pKa = 3.48QWRR43 pKa = 11.84DD44 pKa = 3.16ARR46 pKa = 11.84PVDD49 pKa = 4.84SVRR52 pKa = 11.84TWFRR56 pKa = 11.84TFFSTLRR63 pKa = 11.84MQGASRR69 pKa = 11.84RR70 pKa = 11.84FSLRR74 pKa = 11.84TLLLRR79 pKa = 11.84RR80 pKa = 11.84PTPLRR85 pKa = 11.84VAGMAGGLNLVRR97 pKa = 11.84GLNVARR103 pKa = 11.84GMSRR107 pKa = 11.84KK108 pKa = 9.01PRR110 pKa = 11.84RR111 pKa = 11.84TSTTSTGWFAFARR124 pKa = 4.07
Molecular weight: 14.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2609239 |
29 |
5463 |
300.0 |
32.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.536 ± 0.044 | 0.988 ± 0.009 |
5.526 ± 0.018 | 4.95 ± 0.024 |
3.632 ± 0.016 | 7.952 ± 0.03 |
2.427 ± 0.015 | 4.707 ± 0.019 |
3.002 ± 0.023 | 10.006 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.324 ± 0.014 | 2.909 ± 0.021 |
5.165 ± 0.023 | 3.74 ± 0.017 |
7.256 ± 0.031 | 5.882 ± 0.027 |
5.578 ± 0.025 | 7.647 ± 0.02 |
1.372 ± 0.011 | 2.401 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
